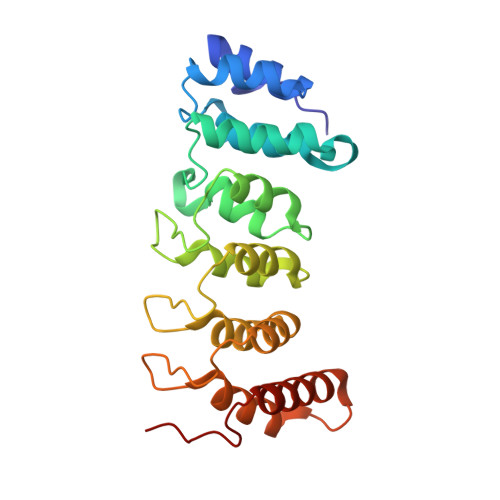
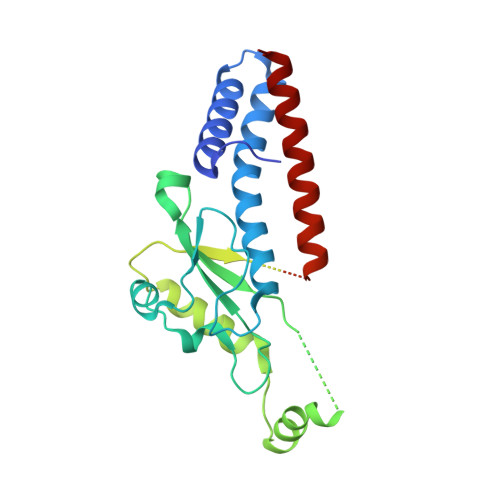
Advanced glycation end-product crosslinking activates a type VI secretion system phospholipase effector protein.
Jensen, S.J., Cuthbert, B.J., Garza-Sanchez, F., Helou, C.C., de Miranda, R., Goulding, C.W., Hayes, C.S.(2024) Nat Commun 15: 8804-8804
- PubMed: 39394186
- DOI: https://doi.org/10.1038/s41467-024-53075-x
- Primary Citation of Related Structures:
7UBZ, 9CYS - PubMed Abstract:
Advanced glycation end-products (AGE) are a pervasive form of protein damage implicated in the pathogenesis of neurodegenerative disease, atherosclerosis and diabetes mellitus. Glycation is typically mediated by reactive dicarbonyl compounds that accumulate in all cells as toxic byproducts of glucose metabolism. Here, we show that AGE crosslinking is harnessed to activate an antibacterial phospholipase effector protein deployed by the type VI secretion system of Enterobacter cloacae. Endogenous methylglyoxal reacts with a specific arginine-lysine pair to tether the N- and C-terminal α-helices of the phospholipase domain. Substitutions at these positions abrogate both crosslinking and toxic phospholipase activity, but in vitro enzyme function can be restored with an engineered disulfide that covalently links the N- and C-termini. Thus, AGE crosslinking serves as a bona fide post-translation modification to stabilize phospholipase structure. Given the ubiquity of methylglyoxal in prokaryotic and eukaryotic cells, these findings suggest that glycation may be exploited more generally to stabilize other proteins. This alternative strategy to fortify tertiary structure could be particularly advantageous in the cytoplasm, where redox potentials preclude disulfide bond formation.
- Department of Molecular, Cellular and Developmental Biology, University of California, Santa Barbara, Santa Barbara, 93106, USA.
Organizational Affiliation: