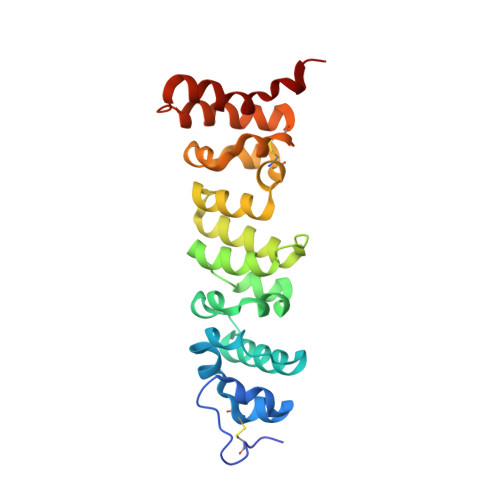
Structures of Cancer Antigen Mesothelin and Its Complexes with Therapeutic Antibodies.
Zhan, J., Lin, D., Watson, N., Esser, L., Tang, W.K., Zhang, A., Liu, X., Hassan, R., Gleinich, A., Shajahan, A., Azadi, P., Pastan, I., Xia, D.(2023) Cancer Res Commun 3: 175-191
- PubMed: 36968141
- DOI: https://doi.org/10.1158/2767-9764.CRC-22-0306
- Primary Citation of Related Structures:
7U9J, 7UED, 8CX3 - PubMed Abstract:
The tumor-associated antigen mesothelin is expressed at high levels on the cell surface of many human cancers, while its expression in normal tissues is limited. The binding of mesothelin to the tumor-associated cancer antigen 125 (CA-125) can lead to heterotypic cell adhesion and tumor metastasis within the pleural and peritoneal cavities. Immunotherapeutic strategies targeting mesothelin are being intensively investigated. Here, we report the crystal structures of mesothelin that reveal a compact, right-handed solenoid consisting of 24 short helices and connecting loops. These helices form a nine-layered spiral coil that resembles ARM/HEAT family proteins. Glycan attachments have been identified in the structure for all three predicted N-glycosylation sites and confirmed with samples from cell culture and patient ascites. The structures of full-length mesothelin and its complex with the Fab of MORAb-009 reveal the interaction of the antibody with the complete epitope, which has not been reported previously. The N-terminal half of mesothelin is conformationally rigid, suitable for eliciting specific antibodies, whereas its C-terminal portion is more flexible. The structure of the C-terminal shedding-resistant fragment of mesothelin complexed with a mAb 15B6 displays an extended linear epitope and helps explain the protection afforded by the antibody for the shedding sites. The structures of full-length mesothelin and its complexes with antibodies reported here are the first to be determined experimentally, providing atomic models for structural organization of this protein and its interactions with antibodies. It offers insights into the function of mesothelin and guidance for further development of therapeutic antibodies.
- Laboratory of Cell Biology, Center for Cancer Research, NCI, Bethesda, Maryland.
Organizational Affiliation: