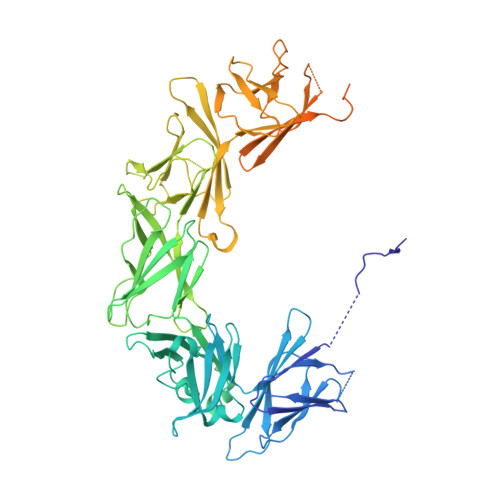
Structural and biochemical insights into lipid transport by VPS13 proteins.
Adlakha, J., Hong, Z., Li, P., Reinisch, K.M.(2022) J Cell Biol 221
- PubMed: 35357422
- DOI: https://doi.org/10.1083/jcb.202202030
- Primary Citation of Related Structures:
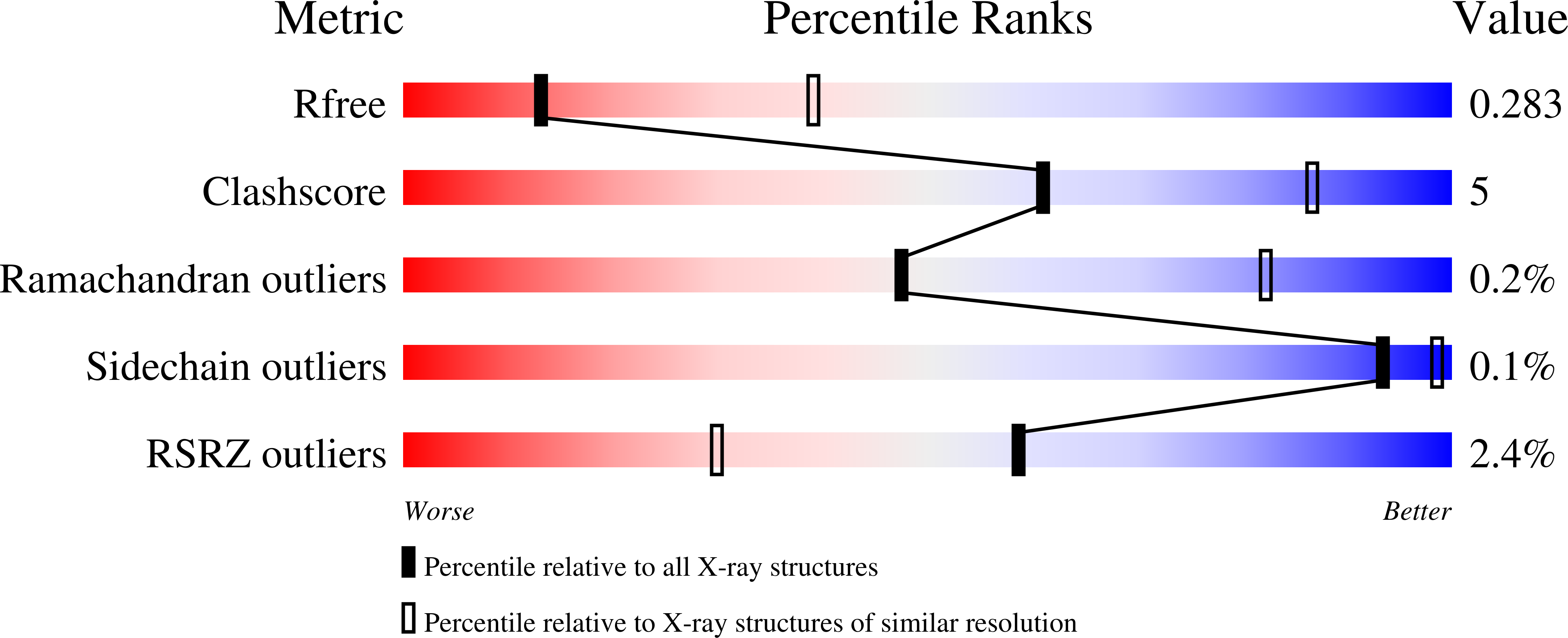
7U8T - PubMed Abstract:
VPS13 proteins are proposed to function at contact sites between organelles as bridges for lipids to move directionally and in bulk between organellar membranes. VPS13s are anchored between membranes via interactions with receptors, including both peripheral and integral membrane proteins. Here we present the crystal structure of VPS13s adaptor binding domain (VAB) complexed with a Pro-X-Pro peptide recognition motif present in one such receptor, the integral membrane protein Mcp1p, and show biochemically that other Pro-X-Pro motifs bind the VAB in the same site. We further demonstrate that Mcp1p and another integral membrane protein that interacts directly with human VPS13A, XK, are scramblases. This finding supports an emerging paradigm of a partnership between bulk lipid transport proteins and scramblases. Scramblases can re-equilibrate lipids between membrane leaflets as lipids are removed from or inserted into the cytosolic leaflet of donor and acceptor organelles, respectively, in the course of protein-mediated transport.
- Department of Cell Biology, Yale School of Medicine, New Haven, CT.
Organizational Affiliation: