Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Bonazzi, S., d'Hennezel, E., Beckwith, R.E.J., Xu, L., Fazal, A., Magracheva, A., Ramesh, R., Cernijenko, A., Antonakos, B., Bhang, H.C., Caro, R.G., Cobb, J.S., Ornelas, E., Ma, X., Wartchow, C.A., Clifton, M.C., Forseth, R.R., Fortnam, B.H., Lu, H., Csibi, A., Tullai, J., Carbonneau, S., Thomsen, N.M., Larrow, J., Chie-Leon, B., Hainzl, D., Gu, Y., Lu, D., Meyer, M.J., Alexander, D., Kinyamu-Akunda, J., Sabatos-Peyton, C.A., Dales, N.A., Zecri, F.J., Jain, R.K., Shulok, J., Wang, Y.K., Briner, K., Porter, J.A., Tallarico, J.A., Engelman, J.A., Dranoff, G., Bradner, J.E., Visser, M., Solomon, J.M.(2023) Cell Chem Biol 30: 235-247.e12
- PubMed: 36863346
- DOI: https://doi.org/10.1016/j.chembiol.2023.02.005
- Primary Citation of Related Structures:
7U8F, 8DEY - PubMed Abstract:
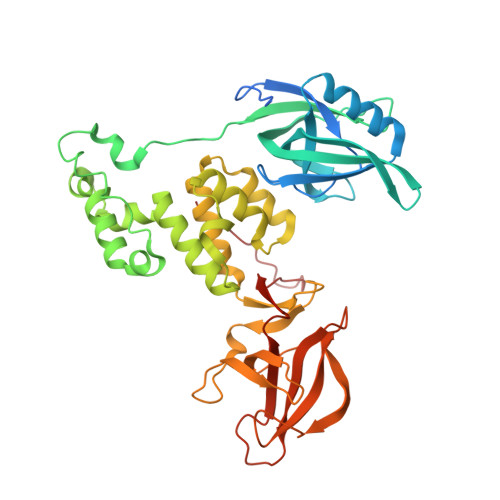
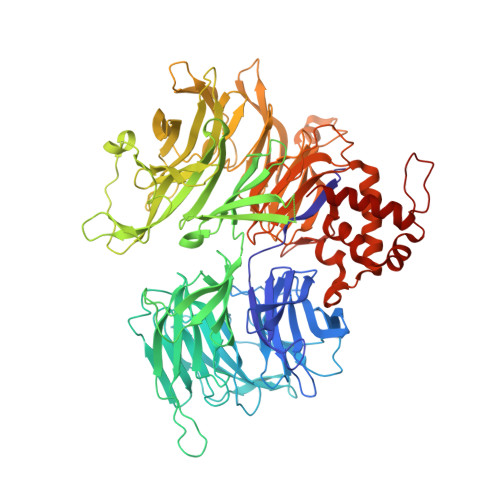
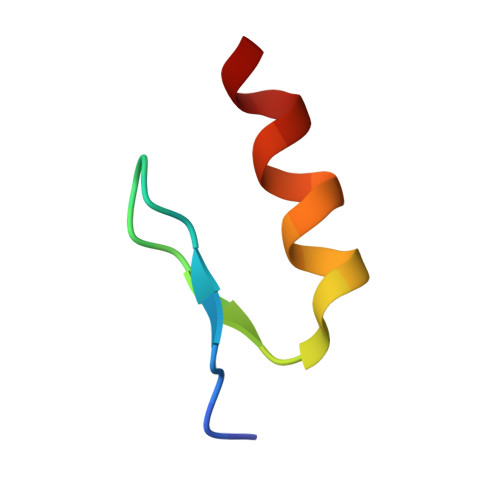
Malignant tumors can evade destruction by the immune system by attracting immune-suppressive regulatory T cells (Treg) cells. The IKZF2 (Helios) transcription factor plays a crucial role in maintaining function and stability of Treg cells, and IKZF2 deficiency reduces tumor growth in mice. Here we report the discovery of NVP-DKY709, a selective molecular glue degrader of IKZF2 that spares IKZF1/3. We describe the recruitment-guided medicinal chemistry campaign leading to NVP-DKY709 that redirected the degradation selectivity of cereblon (CRBN) binders from IKZF1 toward IKZF2. Selectivity of NVP-DKY709 for IKZF2 was rationalized by analyzing the DDB1:CRBN:NVP-DKY709:IKZF2(ZF2 or ZF2-3) ternary complex X-ray structures. Exposure to NVP-DKY709 reduced the suppressive activity of human Treg cells and rescued cytokine production in exhausted T-effector cells. In vivo, treatment with NVP-DKY709 delayed tumor growth in mice with a humanized immune system and enhanced immunization responses in cynomolgus monkeys. NVP-DKY709 is being investigated in the clinic as an immune-enhancing agent for cancer immunotherapy.
- Novartis Institutes for Biomedical Research, Cambridge, MA, USA. Electronic address: simone.bonazzi@novartis.com.
Organizational Affiliation: