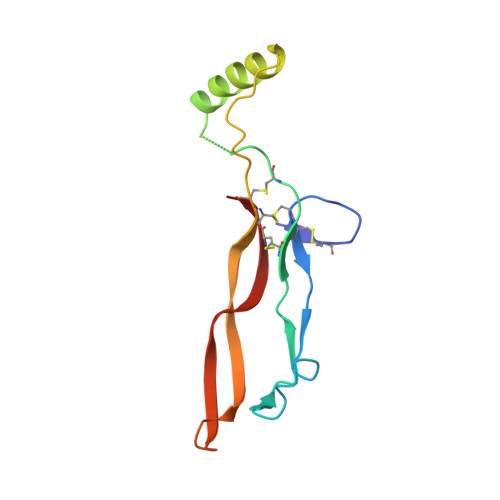
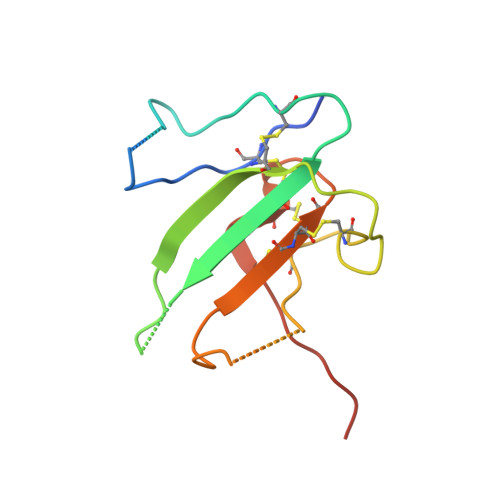
Type II BMP and activin receptors BMPR2 and ACVR2A share a conserved mode of growth factor recognition.
Chu, K.Y., Malik, A., Thamilselvan, V., Martinez-Hackert, E.(2022) J Biological Chem 298: 102076-102076
- PubMed: 35643319
- DOI: https://doi.org/10.1016/j.jbc.2022.102076
- Primary Citation of Related Structures:
7U5O, 7U5P - PubMed Abstract:
BMPR2 is a type II Transforming Growth Factor (TGF)-β family receptor that is fundamentally associated with pulmonary arterial hypertension (PAH) in humans. BMPR2 shares functional similarities with the type II activin receptors ACVR2A and ACVR2B, as it interacts with an overlapping group of TGF-β family growth factors (GFs). However, how BMPR2 recognizes GFs remains poorly understood. Here, we solved crystal structures of BMPR2 in complex with the GF activin B and of ACVR2A in complex with the related GF activin A. We show that both BMPR2 and ACVR2A bind GFs with nearly identical geometry using a conserved hydrophobic hot spot, while differences in contacting residues are predominantly found in loop areas. Upon further exploration of the GF-binding spectrum of the two receptors, we found that although many GFs bind both receptors, the high-affinity BMPR2 GFs comprise BMP15, BMP10, and Nodal, whereas those of ACVR2A are activin A, activin B, and GDF11. Lastly, we evaluated GF-binding domain BMPR2 variants found in human PAH patients. We demonstrate that mutations within the GF-binding interface resulted in loss of GF binding, while mutations in loop areas allowed BMPR2 to retain the ability to bind cognate GFs with high affinity. In conclusion, the in vitro activities of BMPR2 variants and the crystal structures reported here indicate biochemically relevant complexes that explain how some GF-binding domain variants can lead to PAH.
- Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, Michigan, USA.
Organizational Affiliation: