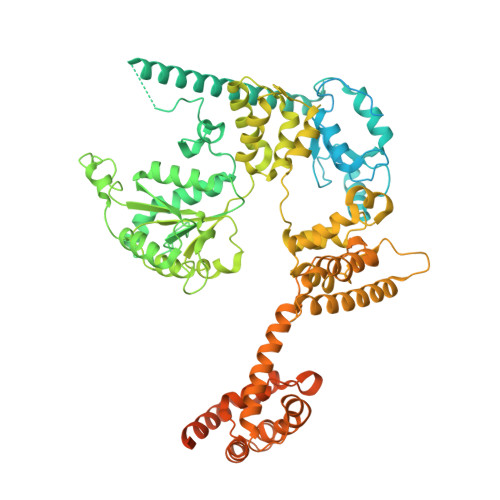
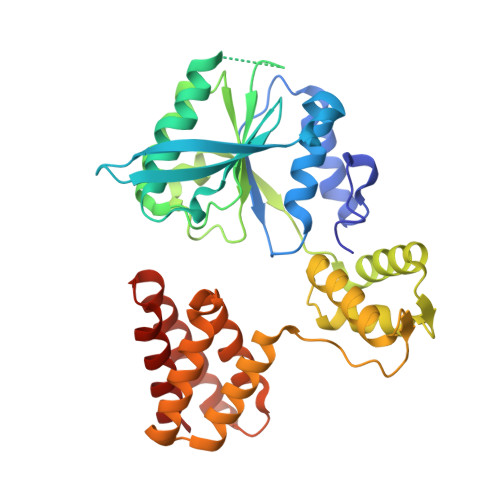
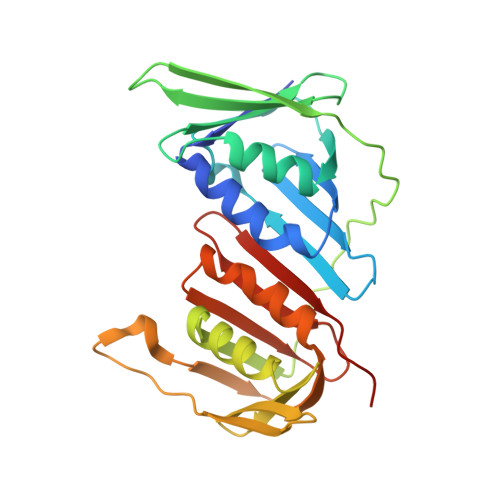
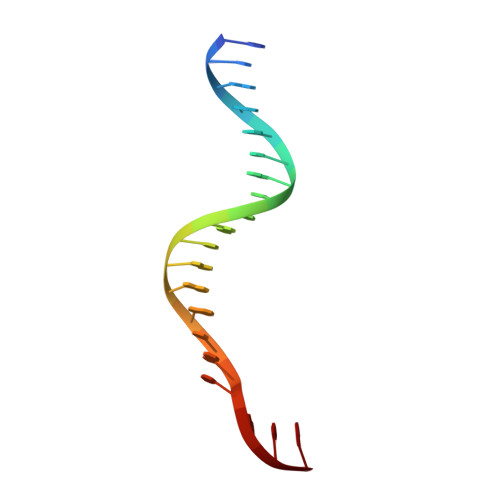
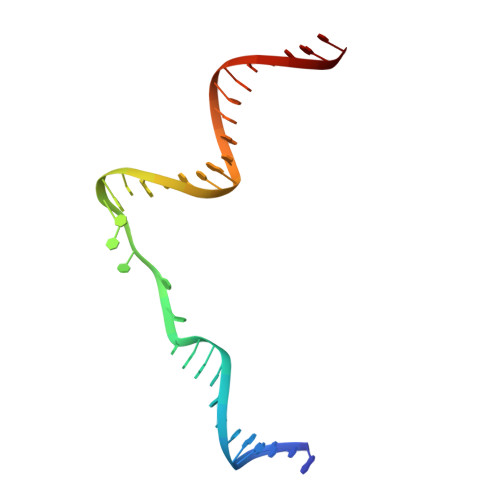
A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA.
Liu, X., Gaubitz, C., Pajak, J., Kelch, B.A.(2022) Elife 11
- PubMed: 35731107
- DOI: https://doi.org/10.7554/eLife.77483
- Primary Citation of Related Structures:
7U19, 7U1A, 7U1P - PubMed Abstract:
Clamp loaders place circular sliding clamp proteins onto DNA so that clamp-binding partner proteins can synthesize, scan, and repair the genome. DNA with nicks or small single-stranded gaps are common clamp-loading targets in DNA repair, yet these substrates would be sterically blocked given the known mechanism for binding of primer-template DNA. Here, we report the discovery of a second DNA binding site in the yeast clamp loader replication factor C (RFC) that aids in binding to nicked or gapped DNA. This DNA binding site is on the external surface and is only accessible in the open conformation of RFC. Initial DNA binding at this site thus provides access to the primary DNA binding site in the central chamber. Furthermore, we identify that this site can partially unwind DNA to create an extended single-stranded gap for DNA binding in RFC's central chamber and subsequent ATPase activation. Finally, we show that deletion of the BRCT domain, a major component of the external DNA binding site, results in defective yeast growth in the presence of DNA damage where nicked or gapped DNA intermediates occur. We propose that RFC's external DNA binding site acts to enhance DNA binding and clamp loading, particularly at DNA architectures typically found in DNA repair.
- Department of Biochemistry and Molecular Biotechnology, University of Massachusetts Chan Medical School, Worcester, United States.
Organizational Affiliation: