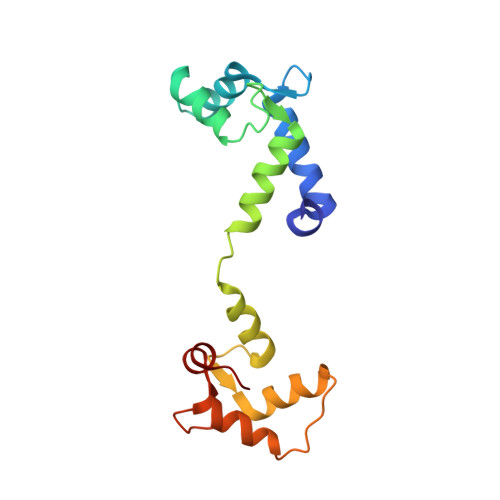
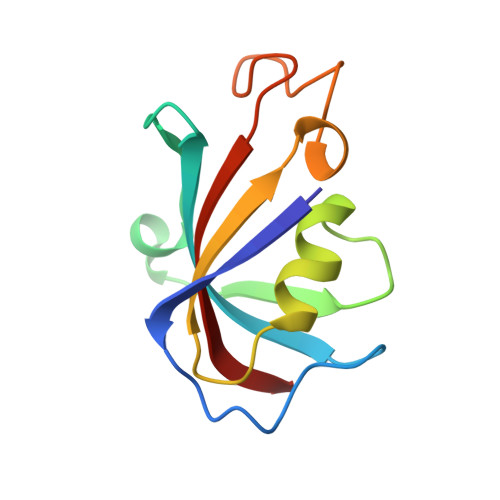
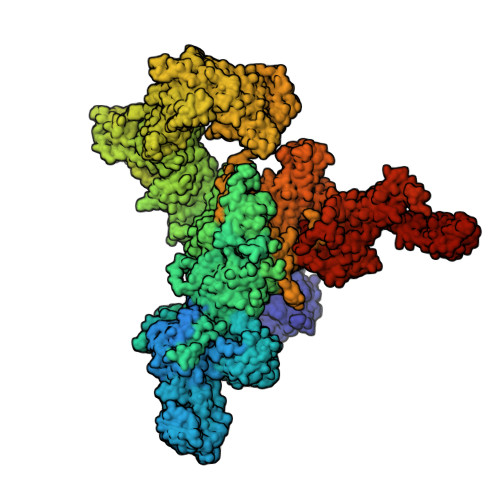
A drug and ATP binding site in type 1 ryanodine receptor.
Melville, Z., Dridi, H., Yuan, Q., Reiken, S., Wronska, A., Liu, Y., Clarke, O.B., Marks, A.R.(2022) Structure 30: 1025
- PubMed: 35580609
- DOI: https://doi.org/10.1016/j.str.2022.04.010
- Primary Citation of Related Structures:
7TZC - PubMed Abstract:
The ryanodine receptor (RyR)/calcium release channel on the sarcoplasmic reticulum (SR) is required for excitation-contraction coupling in skeletal and cardiac muscle. Inherited mutations and stress-induced post-translational modifications result in an SR Ca 2+ leak that causes skeletal myopathies, heart failure, and exercise-induced sudden death. A class of therapeutics known as Rycals prevent the RyR-mediated leak, are effective in preventing disease progression and restoring function in animal models, and are in clinical trials for patients with muscle and heart disorders. Using cryogenic-electron microscopy, we present a model of RyR1 with a 2.45-Å resolution before local refinement, revealing a binding site in the RY1&2 domain (3.10 Å local resolution), where the Rycal ARM210 binds cooperatively with ATP and stabilizes the closed state of RyR1.
- Department of Physiology and Cellular Biophysics, Columbia University, New York, NY, USA.
Organizational Affiliation: