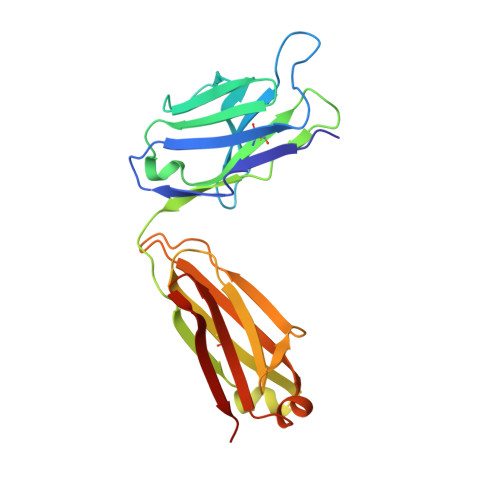
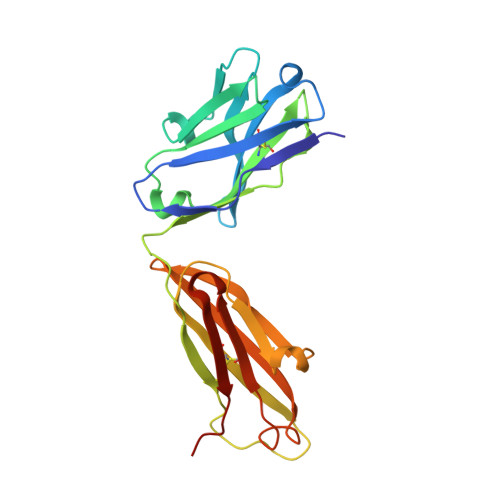
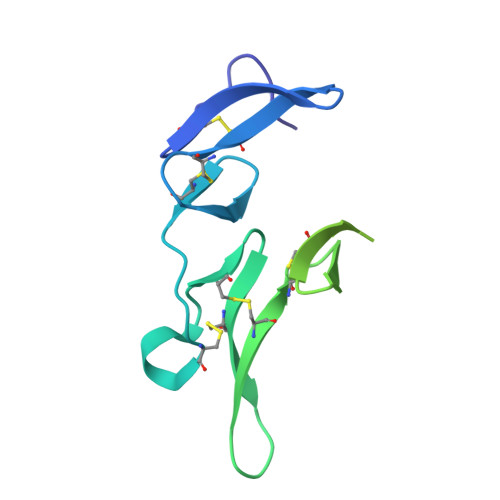
Structural and immunological differences in Plasmodium falciparum sexual stage transmission-blocking vaccines comprised of Pfs25-EPA nanoparticles.
MacDonald, N.J., Singh, K., Reiter, K., Nguyen, V., Shimp Jr., R., Gittis, A.G., Chen, B., Burkhardt, M., Zhang, B., Wang, Z., Herrera, R., Moler, M., Lee, D.Y., Orr-Gonzalez, S., Herrod, J., Lambert, L.E., Rausch, K.M., Muratova, O., Jones, D.S., Wu, Y., Jin, A.J., Garboczi, D.N., Duffy, P.E., Narum, D.L.(2023) NPJ Vaccines 8: 56-56
- PubMed: 37061547
- DOI: https://doi.org/10.1038/s41541-023-00655-5
- Primary Citation of Related Structures:
7TXW - PubMed Abstract:
Development of a malaria vaccine that blocks transmission of different parasite stages to humans and mosquitoes is considered critical for elimination efforts. A vaccine using Pfs25, a protein on the surface of zygotes and ookinetes, is under investigation as a transmission-blocking vaccine (TBV) that would interrupt parasite passage from mosquitoes to humans. The most extensively studied Pfs25 TBVs use Pichia pastoris-produced recombinant forms of Pfs25, chemically conjugated to a recombinant carrier protein, ExoProtein A (EPA). The recombinant form of Pfs25 first used in humans was identified as Pfs25H, which contained a total of 14 heterologous amino acid residues located at the amino- and carboxyl-termini including a His6 affinity tag. A second recombinant Pfs25, identified as Pfs25M, was produced to remove the heterologous amino acid residues and conjugated to EPA (Pfs25M-EPA). Here, monomeric Pfs25M was characterized biochemically and biophysically for identity, purity, and integrity including protein structure to assess its comparability with Pfs25H. Although the biological activities of Pfs25H and Pfs25M, whether generated by monomeric forms or conjugated nanoparticles, appeared similar, fine-mapping studies with two transmission-blocking monoclonal antibodies detected structural and immunological differences. In addition, evaluation of antisera generated against conjugated Pfs25H or Pfs25M nanoparticles in nonhuman primates identified polyclonal IgG that recognized these structural differences.
- Laboratory of Malaria Immunology and Vaccinology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, 29 Lincoln Drive, Bethesda, MD, 20892, USA.
Organizational Affiliation: