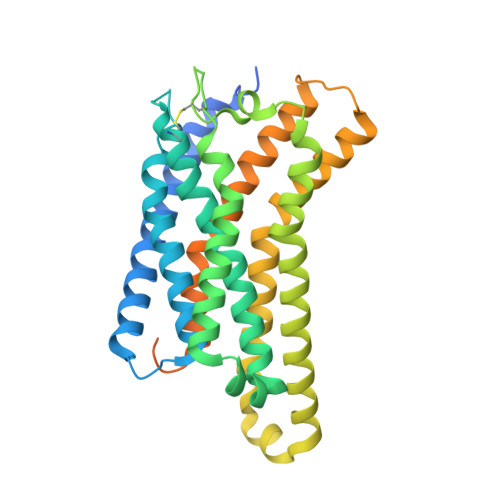
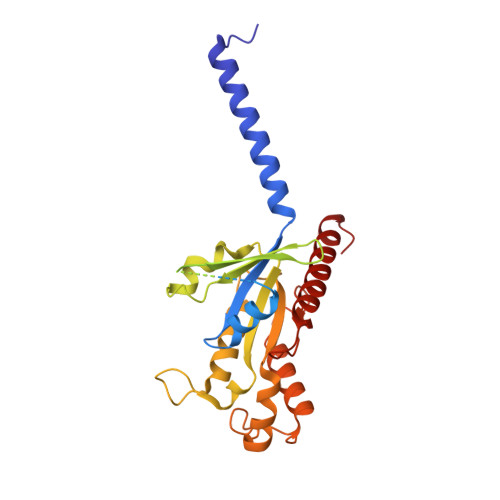
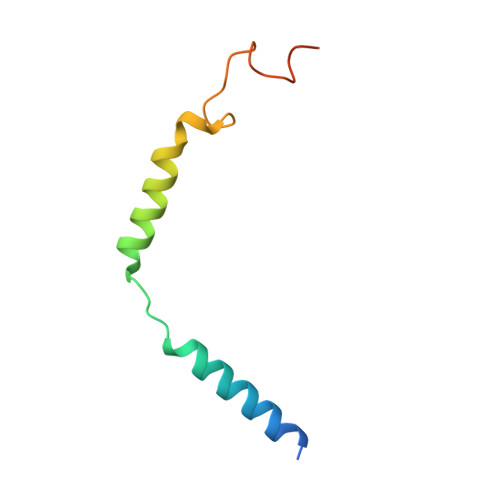
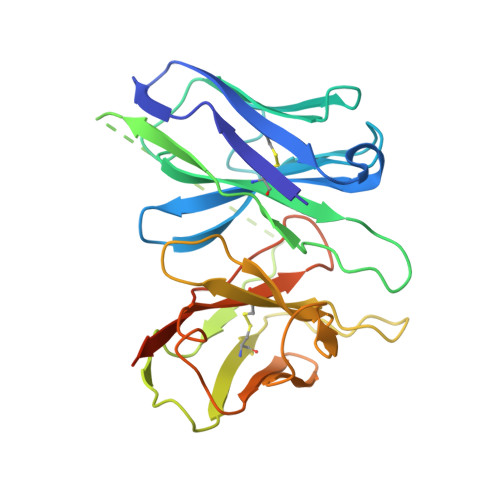
Structures of oxysterol sensor EBI2/GPR183, a key regulator of the immune response.
Chen, H., Huang, W., Li, X.(2022) Structure 30: 1016
- PubMed: 35537452
- DOI: https://doi.org/10.1016/j.str.2022.04.006
- Primary Citation of Related Structures:
7TUY, 7TUZ - PubMed Abstract:
Oxysterols induce the migration of B-lymphocytes and dendritic cells to interfollicular regions of lymphoid tissues through binding the EBI2 (GPR183) to stimulate effective adaptive immunity and antibody production during infection. Aberrant EBI2 signaling is implicated in inflammatory bowel disease, sclerosis, and infectious disease. Here, we report the cryo-EM structures of an EBI2-G i signaling complex with its endogenous agonist 7α,25-OHC and that of an inactive EBI2 bound to the inverse agonist GSK682753A. These structures reveal an agonist binding site for the oxysterol and a potential ligand entrance site exposed to the lipid bilayer. Mutations within the oxysterol binding site and the Gα i interface attenuate G protein signaling and abolish oxysterol-mediated cell migration indicating that G protein signaling directly involves in the oxysterol-EBI2 pathway. Together, these findings provide new insight into how EBI2 is activated by an oxysterol ligand and will facilitate the development of therapeutic approaches that target EBI2-linked diseases.
- Department of Molecular Genetics, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.
Organizational Affiliation: