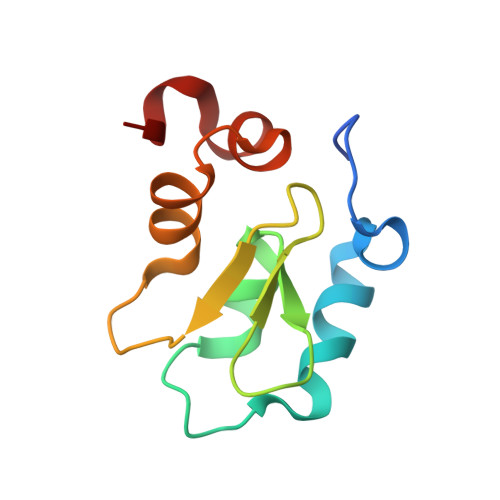
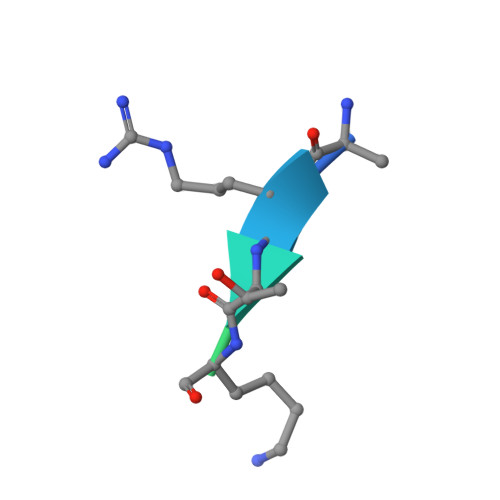
Molecular basis for nuclear accumulation and targeting of the inhibitor of apoptosis BIRC2.
Tencer, A.H., Yu, Y., Causse, S.Z., Campbell, G.R., Klein, B.J., Xuan, H., Cartier, J., Miles, M.A., Gaurav, N., Zadoroznyj, A., Holt, T.A., Wen, H., Hawkins, C.J., Spector, S.A., Dubrez, L., Shi, X., Kutateladze, T.G.(2023) Nat Struct Mol Biol 30: 1265-1274
- PubMed: 37524969
- DOI: https://doi.org/10.1038/s41594-023-01044-1
- Primary Citation of Related Structures:
7TRL, 7TRM - PubMed Abstract:
The inhibitor of apoptosis protein BIRC2 regulates fundamental cell death and survival signaling pathways. Here we show that BIRC2 accumulates in the nucleus via binding of its second and third BIR domains, BIRC2 BIR2 and BIRC2 BIR3 , to the histone H3 tail and report the structure of the BIRC2 BIR3 -H3 complex. RNA-seq analysis reveals that the genes involved in interferon and defense response signaling and cell-cycle regulation are most affected by depletion of BIRC2. Overexpression of BIRC2 delays DNA damage repair and recovery of the cell-cycle progression. We describe the structural mechanism for targeting of BIRC2 BIR3 by a potent but biochemically uncharacterized small molecule inhibitor LCL161 and demonstrate that LCL161 disrupts the association of endogenous BIRC2 with H3 and stimulates cell death in cancer cells. We further show that LCL161 mediates degradation of BIRC2 in human immunodeficiency virus type 1-infected human CD4 + T cells. Our findings provide mechanistic insights into the nuclear accumulation of and blocking BIRC2.
- Department of Pharmacology, University of Colorado School of Medicine, Aurora, CO, USA.
Organizational Affiliation: