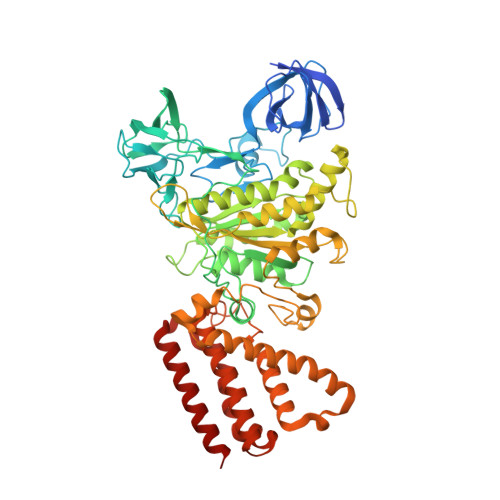
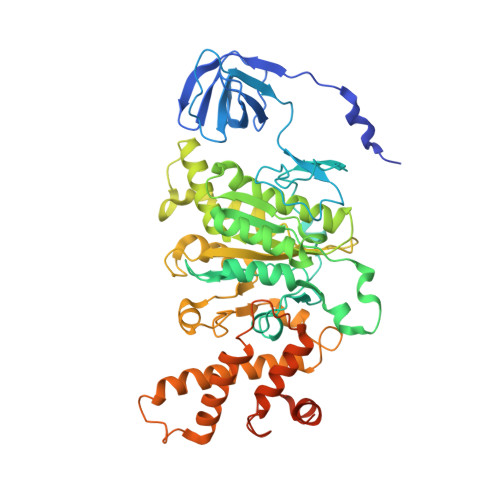
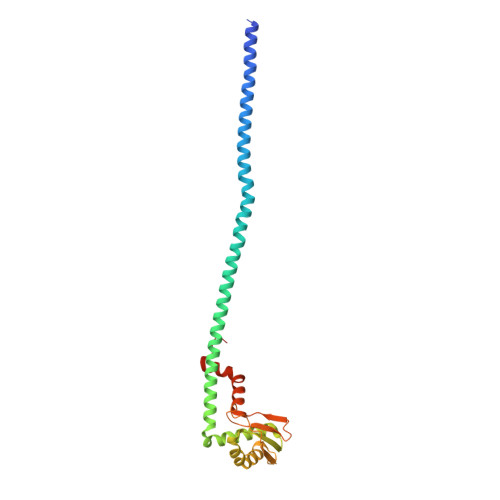
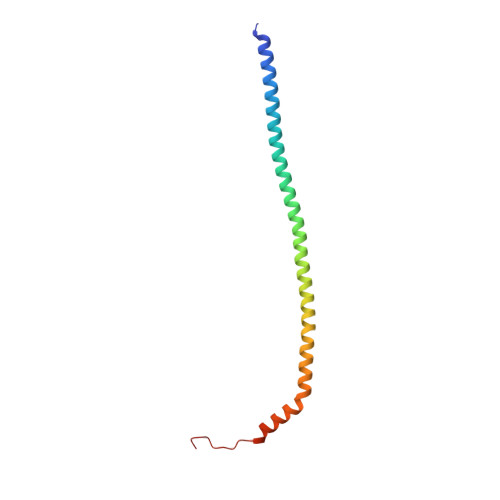
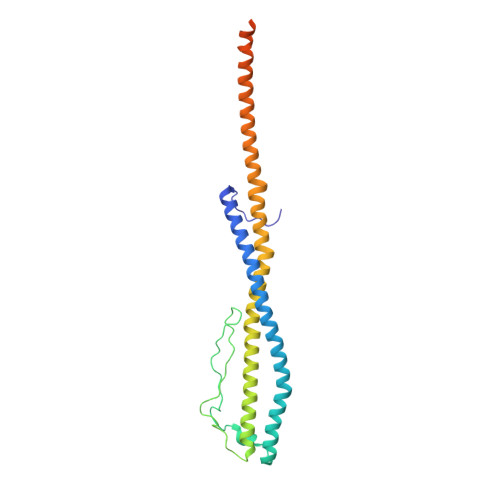
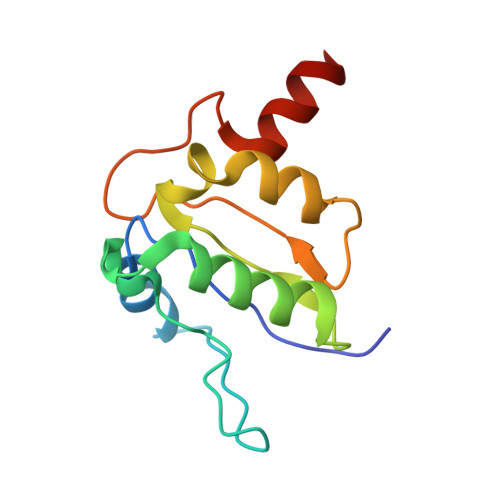
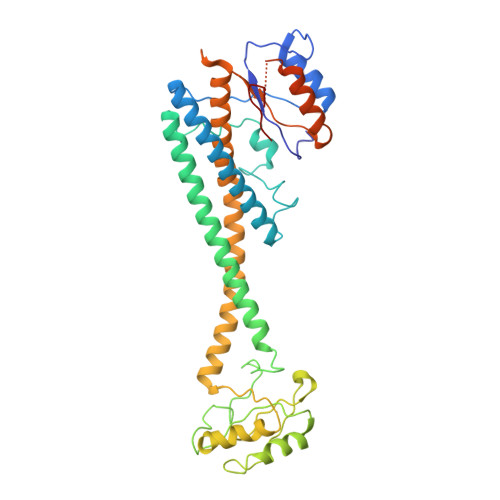
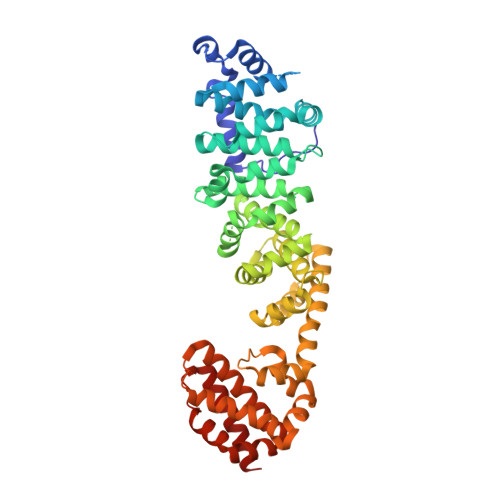
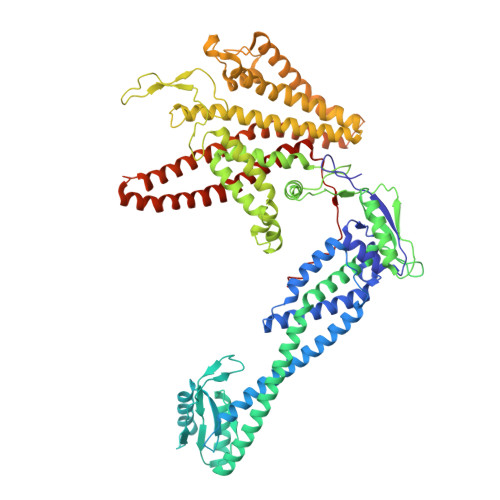
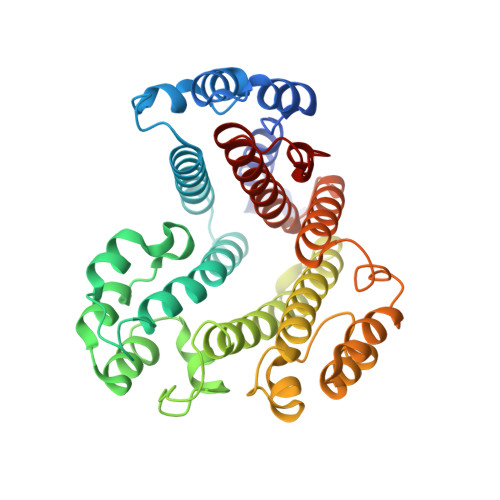
Coordinated conformational changes in the V 1 complex during V-ATPase reversible dissociation.
Vasanthakumar, T., Keon, K.A., Bueler, S.A., Jaskolka, M.C., Rubinstein, J.L.(2022) Nat Struct Mol Biol 29: 430-439
- PubMed: 35469063
- DOI: https://doi.org/10.1038/s41594-022-00757-z
- Primary Citation of Related Structures:
7TMM, 7TMO, 7TMP, 7TMQ, 7TMR, 7TMS, 7TMT - PubMed Abstract:
Vacuolar-type ATPases (V-ATPases) are rotary enzymes that acidify intracellular compartments in eukaryotic cells. These multi-subunit complexes consist of a cytoplasmic V 1 region that hydrolyzes ATP and a membrane-embedded V O region that transports protons. V-ATPase activity is regulated by reversible dissociation of the two regions, with the isolated V 1 and V O complexes becoming autoinhibited on disassembly and subunit C subsequently detaching from V 1 . In yeast, assembly of the V 1 and V O regions is mediated by the regulator of the ATPase of vacuoles and endosomes (RAVE) complex through an unknown mechanism. We used cryogenic-electron microscopy of yeast V-ATPase to determine structures of the intact enzyme, the dissociated but complete V 1 complex and the V 1 complex lacking subunit C. On separation, V 1 undergoes a dramatic conformational rearrangement, with its rotational state becoming incompatible for reassembly with V O . Loss of subunit C allows V 1 to match the rotational state of V O , suggesting how RAVE could reassemble V 1 and V O by recruiting subunit C.
- Molecular Medicine Program, The Hospital for Sick Children, Toronto, Ontario, Canada.
Organizational Affiliation: