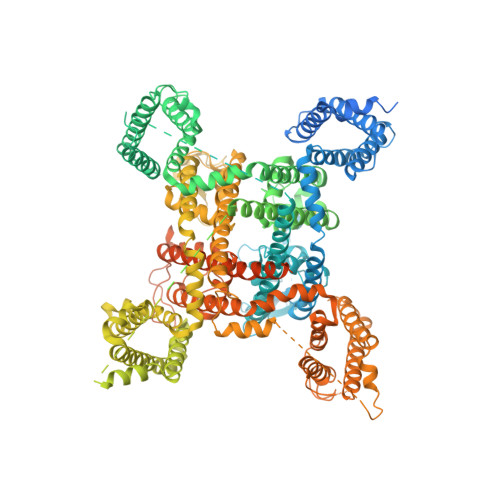
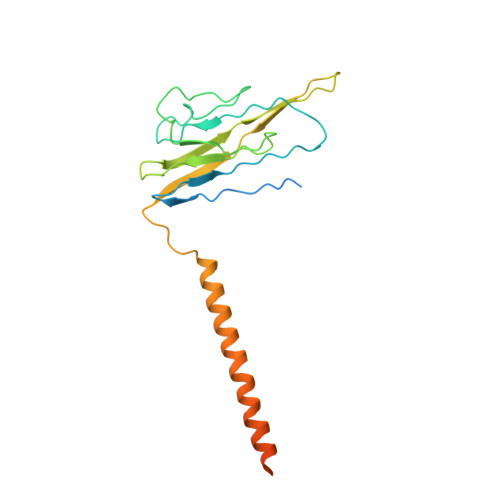
Structure-guided unlocking of Na X reveals a non-selective tetrodotoxin-sensitive cation channel.
Noland, C.L., Chua, H.C., Kschonsak, M., Heusser, S.A., Braun, N., Chang, T., Tam, C., Tang, J., Arthur, C.P., Ciferri, C., Pless, S.A., Payandeh, J.(2022) Nat Commun 13: 1416-1416
- PubMed: 35301303
- DOI: https://doi.org/10.1038/s41467-022-28984-4
- Primary Citation of Related Structures:
7TJ8, 7TJ9 - PubMed Abstract:
Unlike classical voltage-gated sodium (Na V ) channels, Na X has been characterized as a voltage-insensitive, tetrodotoxin-resistant, sodium (Na + )-activated channel involved in regulating Na + homeostasis. However, Na X remains refractory to functional characterization in traditional heterologous systems. Here, to gain insight into its atypical physiology, we determine structures of the human Na X channel in complex with the auxiliary β3-subunit. Na X reveals structural alterations within the selectivity filter, voltage sensor-like domains, and pore module. We do not identify an extracellular Na + -sensor or any evidence for a Na + -based activation mechanism in Na X . Instead, the S6-gate remains closed, membrane lipids fill the central cavity, and the domain III-IV linker restricts S6-dilation. We use protein engineering to identify three pore-wetting mutations targeting the hydrophobic S6-gate that unlock a robust voltage-insensitive leak conductance. This constitutively active Na X -QTT channel construct is non-selective among monovalent cations, inhibited by extracellular calcium, and sensitive to classical Na V channel blockers, including tetrodotoxin. Our findings highlight a functional diversity across the Na V channel scaffold, reshape our understanding of Na X physiology, and provide a template to demystify recalcitrant ion channels.
- Department of Structural Biology, Genentech Inc., South San Francisco, CA, 94080, USA.
Organizational Affiliation: