Structural Analyses of CrtJ and Its B 12 -Binding Co-Regulators SAerR and LAerR from the Purple Photosynthetic Bacterium Rhodobacter capsulatus.
Dragnea, V., Gonzalez-Gutierrez, G., Bauer, C.E.(2022) Microorganisms 10
- PubMed: 35630357
- DOI: https://doi.org/10.3390/microorganisms10050912
- Primary Citation of Related Structures:
7TE2 - PubMed Abstract:
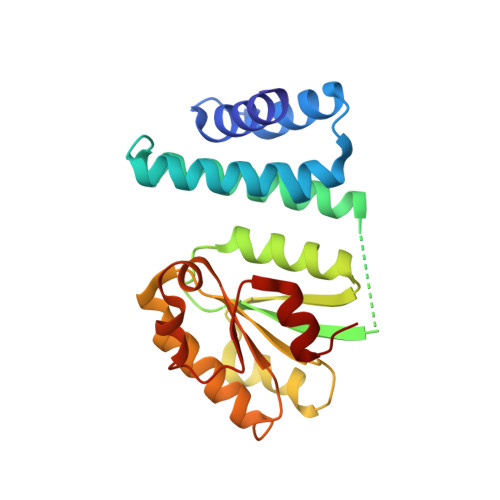
Among purple photosynthetic bacteria, the transcription factor CrtJ is a major regulator of photosystem gene expression. Depending on growing conditions, CrtJ can function as an aerobic repressor or an anaerobic activator of photosystem genes. Recently, CrtJ's activity was shown to be modulated by two size variants of a B 12 binding co-regulator called SAerR and LAerR in Rhodobacter capsulatus . The short form, SAerR, promotes CrtJ repression, while the longer variant, LAerR, converts CrtJ into an activator. In this study, we solved the crystal structure of R. capsulatus SAerR at a 2.25 Å resolution. Hydroxycobalamin bound to SAerR is sandwiched between a 4-helix bundle cap, and a Rossman fold. This structure is similar to a AerR-like domain present in CarH from Thermus termophilus , which is a combined photoreceptor/transcription regulator. We also utilized AlphaFold software to predict structures for the LAerR, CrtJ, SAerR-CrtJ and LAerR-CrtJ co-complexes. These structures provide insights into the role of B 12 and an LAerR N-terminal extension in regulating the activity of CrtJ.
- Molecular and Cellular Biochemistry Department, Indiana University, Bloomington, IN 47405, USA.
Organizational Affiliation: