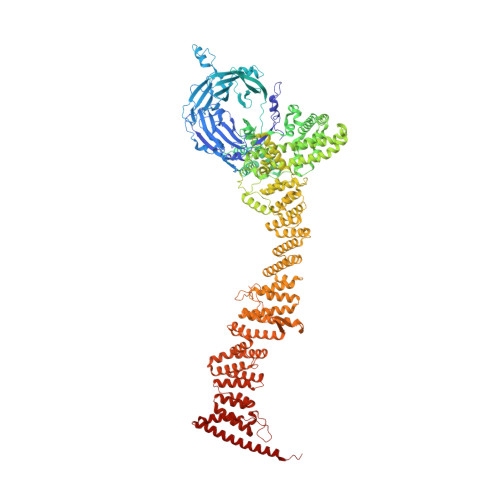
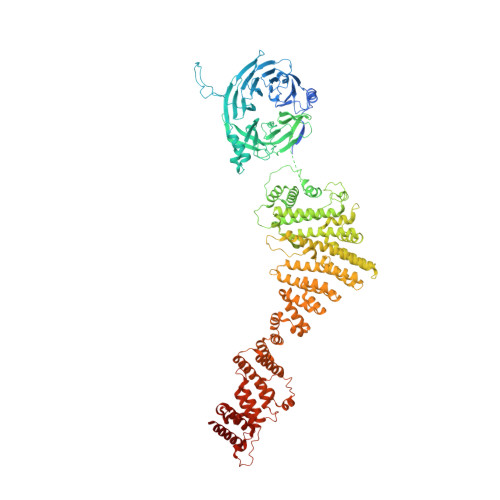
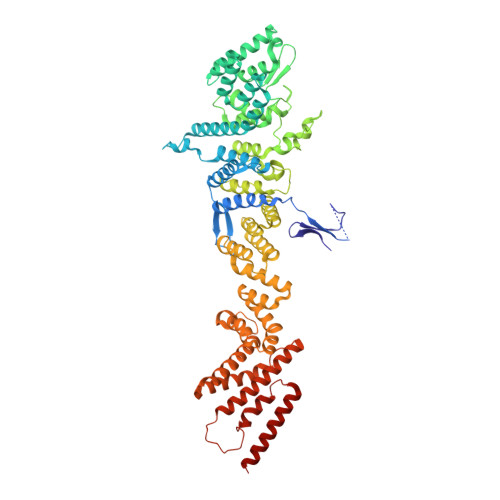
Structure of cytoplasmic ring of nuclear pore complex by integrative cryo-EM and AlphaFold.
Fontana, P., Dong, Y., Pi, X., Tong, A.B., Hecksel, C.W., Wang, L., Fu, T.M., Bustamante, C., Wu, H.(2022) Science 376: eabm9326-eabm9326
- PubMed: 35679401
- DOI: https://doi.org/10.1126/science.abm9326
- Primary Citation of Related Structures:
7TDZ - PubMed Abstract:
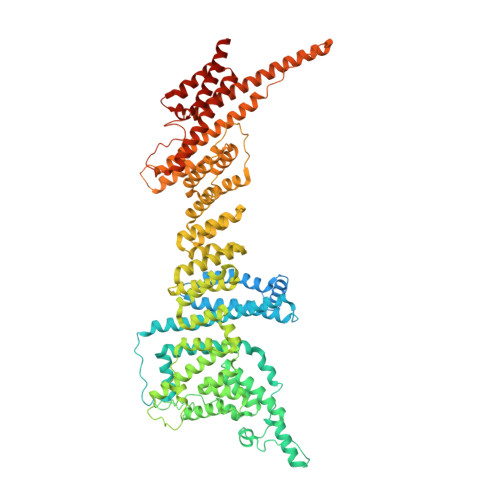
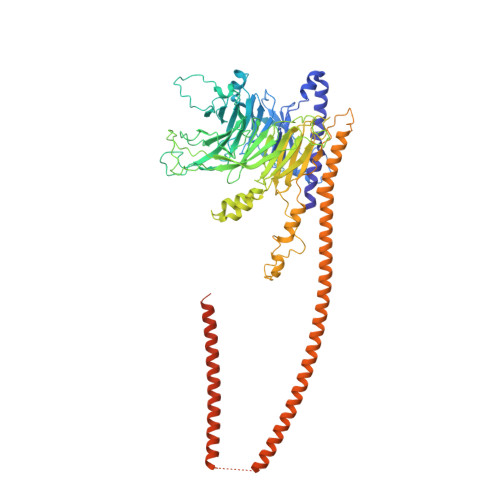
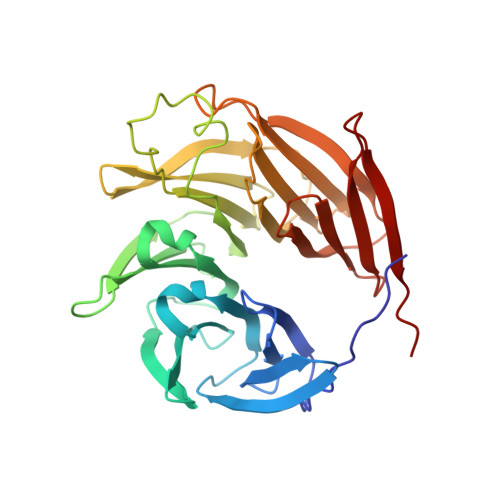
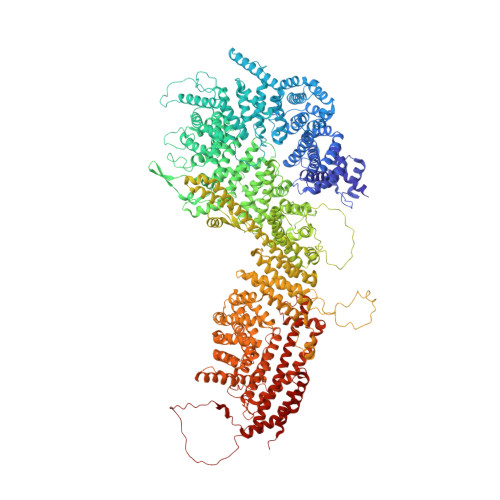
INTRODUCTION The nuclear pore complex (NPC) is the molecular conduit in the nuclear membrane of eukaryotic cells that regulates import and export of biomolecules between the nucleus and the cytosol, with vertebrate NPCs ~110 to 125 MDa in molecular mass and ~120 nm in diameter. NPCs are organized into four main rings: the cytoplasmic ring (CR) at the cytosolic side, the inner ring and the luminal ring on the plane of the nuclear membrane, and the nuclear ring facing the nucleus. Each ring possesses an approximate eightfold symmetry and is composed of multiple copies of different nucleoporins. NPCs have been implicated in numerous biological processes, and their dysfunctions are associated with a growing number of serious human diseases. However, despite pioneering studies from many groups over the past two decades, we still lack a full understanding of NPCs' organization, dynamics, and complexity. RATIONALE We used the Xenopus laevis oocyte as a model system for the structural characterization because each oocyte possesses a large number of NPC particles that can be visualized on native nuclear membranes without the aid of detergent extraction. We used single-particle cryo-electron microscopy (cryo-EM) analysis on data collected at different stage tilt angles for three-dimensional reconstruction and structure prediction with AlphaFold for model building. RESULTS We reconstructed the CR map of X. laevis NPC at 6.9 and 6.7 Å resolutions for the full CR protomer and a core region, respectively, and predicted the structures of the individual nucleoporins using AlphaFold because no high-resolution models of X. laevis Nups were available. For any ambiguous subunit interactions, we also predicted complex structures, which further guided model fitting of the CR protomer. We placed the nucleoporin or complex structures into the CR density to obtain an almost full CR atomic model, composed of the inner and outer Y-complexes, two copies of Nup205, two copies of the Nup214-Nup88-Nup62 complex, one Nup155, and five copies of Nup358. In particular, we predicted the largest protein in the NPC, Nup358, as having an S-shaped globular domain, a coiled-coil domain, and a largely disordered C-terminal region containing phenylalanine-glycine (FG) repeats previously shown to form a gel-like condensate phase for selective cargo passage. Four of the Nup358 copies clamp around the inner and outer Y-complexes to stabilize the CR, and the fifth Nup358 situates in the center of the cluster of clamps. AlphaFold also predicted a homo-oligomeric, likely specifically pentameric, coiled-coil structure of Nup358 that may provide the avidity for Nup358 recruitment to the NPC and for lowering the threshold for Nup358 condensation in NPC biogenesis. CONCLUSION Our studies offer an example of integrative cryo-EM and structure prediction as a general approach for attaining more precise models of megadalton protein complexes from medium-resolution density maps. The more accurate and almost complete model of the CR presented here expands our understanding of the molecular interactions in the NPC and represents a substantial step forward toward the molecular architecture of a full NPC, with implications for NPC function, biogenesis, and regulation. [Figure: see text].
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: