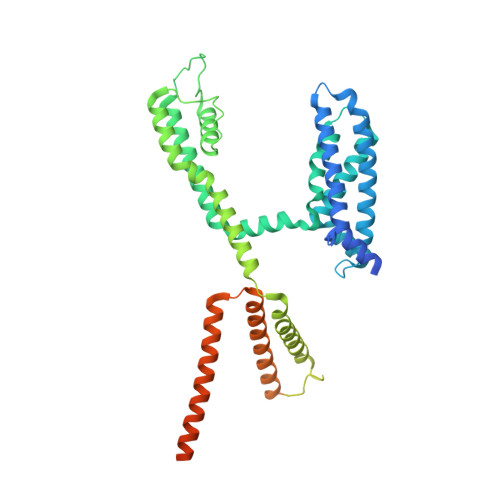
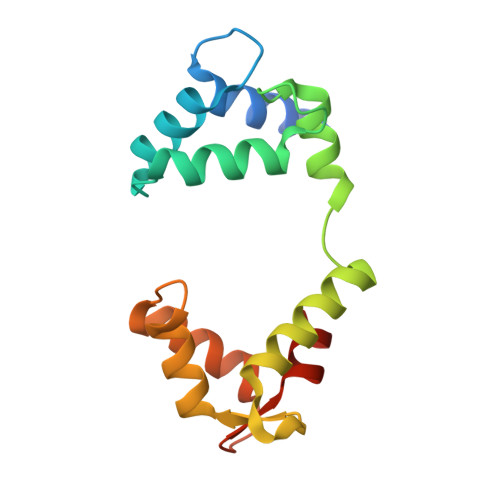
Structural and electrophysiological basis for the modulation of KCNQ1 channel currents by ML277.
Willegems, K., Eldstrom, J., Kyriakis, E., Ataei, F., Sahakyan, H., Dou, Y., Russo, S., Van Petegem, F., Fedida, D.(2022) Nat Commun 13: 3760-3760
- PubMed: 35768468
- DOI: https://doi.org/10.1038/s41467-022-31526-7
- Primary Citation of Related Structures:
7TCI, 7TCP - PubMed Abstract:
The KCNQ1 ion channel plays critical physiological roles in electrical excitability and K + recycling in organs including the heart, brain, and gut. Loss of function is relatively common and can cause sudden arrhythmic death, sudden infant death, epilepsy and deafness. Here, we report cryogenic electron microscopic (cryo-EM) structures of Xenopus KCNQ1 bound to Ca 2+ /Calmodulin, with and without the KCNQ1 channel activator, ML277. A single binding site for ML277 was identified, localized to a pocket lined by the S4-S5 linker, S5 and S6 helices of two separate subunits. Several pocket residues are not conserved in other KCNQ isoforms, explaining specificity. MD simulations and point mutations support this binding location for ML277 in open and closed channels and reveal that prevention of inactivation is an important component of the activator effect. Our work provides direction for therapeutic intervention targeting KCNQ1 loss of function pathologies including long QT interval syndrome and seizures.
- Department of Anesthesiology, Pharmacology and Therapeutics, University of British Columbia, Vancouver, BC, Canada.
Organizational Affiliation: