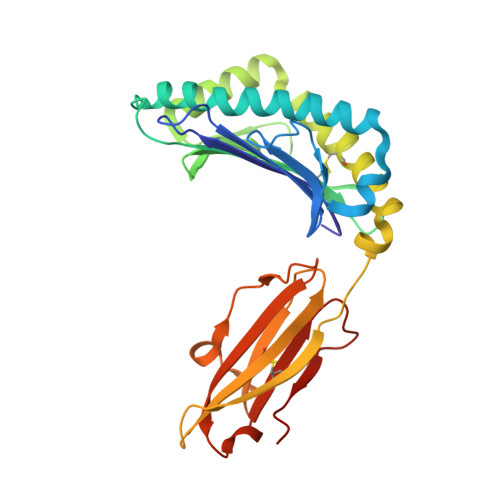
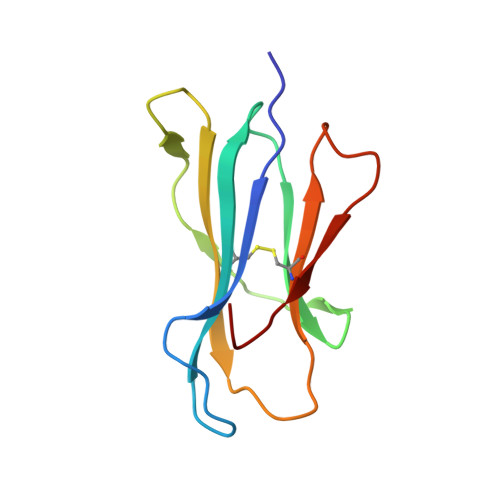
Cutting Edge: Unconventional CD8 + T Cell Recognition of a Naturally Occurring HLA-A*02:01-Restricted 20mer Epitope.
Meeuwsen, M.H., Wouters, A.K., Hagedoorn, R.S., Kester, M.G.D., Remst, D.F.G., van der Steen, D.M., de Ru, A., van Veelen, P.A., Rossjohn, J., Gras, S., Falkenburg, J.H.F., Heemskerk, M.H.M.(2022) J Immunol 208: 1851-1856
- PubMed: 35379743
- DOI: https://doi.org/10.4049/jimmunol.2101208
- Primary Citation of Related Structures:
7T5M - PubMed Abstract:
Unconventional HLA class I-restricted CD8 + T cell epitopes, longer than 10 aa, have been implicated to play a role in human immunity against viruses and cancer. T cell recognition of long peptides, centrally bulging from the HLA cleft, has been described previously. Alternatively, long peptides can contain a linear HLA-bound core peptide, with a N- or C-terminal peptide "tail" extending from the HLA peptide binding groove. The role of such a peptide "tail" in CD8 + T cell recognition remains unclear. In this study, we identified a 20mer peptide (FLPTPEELGLLGPPRPQVLA [FLP]) derived from the IL-27R subunit α gene restricted to HLA-A*02:01, for which we solved the crystal structure and demonstrated a long C-terminal "tail" extension. FLP-specific T cell clones demonstrated various recognition modes, some T cells recognized the FLP core peptide, while for other T cells the peptide tail was essential for recognition. These results demonstrate a crucial role for a C-terminal peptide tail in immunogenicity.
- Department of Hematology, Leiden University Medical Center, Leiden, the Netherlands; m.h.meeuwsen@lumc.nl m.h.m.heemskerk@lumc.nl.
Organizational Affiliation: