The Structural Basis for Recognition of Human Leukocyte Antigen Class I Molecules by the Pan-HLA Antibody W6/32.
Pymm, P., Saunders, P.M., Anand, S., MacLachlan, B.J., Faoro, C., Hitchen, C., Rossjohn, J., Brooks, A.G., Vivian, J.P.(2024) J Immunol 213: 876-885
- PubMed: 39093013
- DOI: https://doi.org/10.4049/jimmunol.2400328
- Primary Citation of Related Structures:
7T0L - PubMed Abstract:
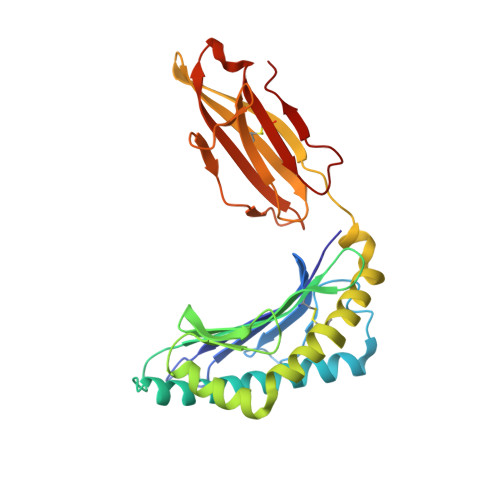
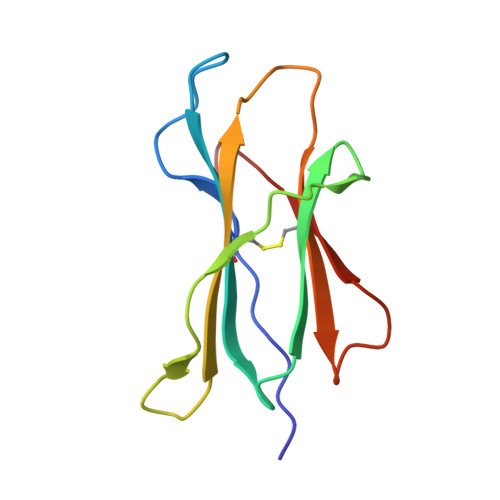
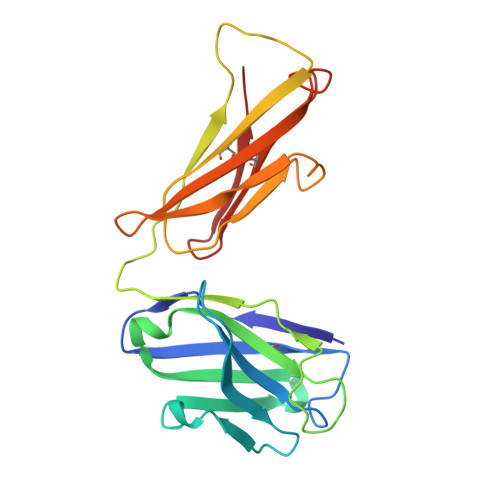
The central immunological role of HLA class I (HLA-I) in presenting peptide Ags to cellular components of the immune system has been the focus of intense study for >60 y. A confounding factor in the study of HLA-I has been the extreme polymorphism of these molecules. The mAb W6/32 has been a fundamental reagent bypassing the issue of polymorphism by recognizing an epitope that is conserved across diverse HLA-I allotypes. However, despite the widespread use of W6/32, the epitope of this Ab has not been definitively mapped. In this study, we present the crystal structure of the Fab fragment of W6/32 in complex with peptide-HLA-B*27:05. W6/32 bound to HLA-B*27:05 beneath the Ag-binding groove, recognizing a discontinuous epitope comprised of the α1, α2, and α3 domains of HLA-I and β2-microglobulin. The epitope comprises a region of low polymorphism reflecting the pan-HLA-I nature of the binding. Notably, the W6/32 epitope neither overlaps the HLA-I binding sites of either T cell Ag receptors or killer cell Ig-like receptors. However, it does coincide with the binding sites for leukocyte Ig-like receptors and CD8 coreceptors. Consistent with this, the use of W6/32 to block the interaction of NK cells with HLA-I only weakly impaired inhibition mediated by KIR3DL1, but impacted HLA-LILR recognition.
- Infection and Immunity Program, Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Victoria, Australia.
Organizational Affiliation: