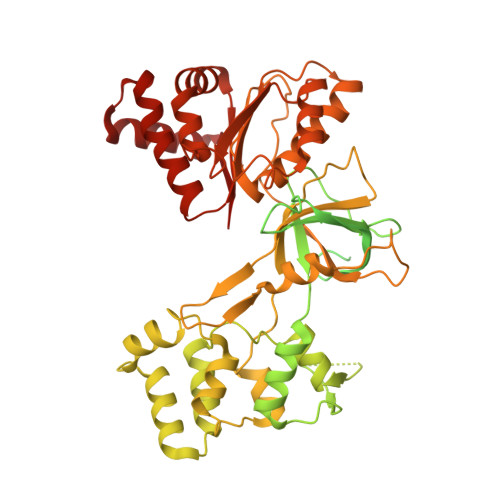
Functional and structural insight into the flexibility of cytochrome P450 reductases from Sorghum bicolor and its implications for lignin composition.
Zhang, B., Munske, G.R., Timokhin, V.I., Ralph, J., Davydov, D.R., Vermerris, W., Sattler, S.E., Kang, C.(2022) J Biological Chem 298: 101761-101761
- PubMed: 35202651
- DOI: https://doi.org/10.1016/j.jbc.2022.101761
- Primary Citation of Related Structures:
7SUX, 7SUZ, 7SV0 - PubMed Abstract:
Plant NADPH-dependent cytochrome P450 reductase (CPR) is a multidomain enzyme that donates electrons for hydroxylation reactions catalyzed by class II cytochrome P450 monooxygenases involved in the synthesis of many primary and secondary metabolites. These P450 enzymes include trans-cinnamate-4-hydroxylase, p-coumarate-3'-hydroxylase, and ferulate-5-hydroxylase involved in monolignol biosynthesis. Because of its role in monolignol biosynthesis, alterations in CPR activity could change the composition and overall output of lignin. Therefore, to understand the structure and function of three CPR subunits from sorghum, recombinant subunits SbCPR2a, SbCPR2b, and SbCPR2c were subjected to X-ray crystallography and kinetic assays. Steady-state kinetic analyses demonstrated that all three CPR subunits supported the oxidation reactions catalyzed by SbC4H1 (CYP73A33) and SbC3'H (CYP98A1). Furthermore, comparing the SbCPR2b structure with the well-investigated CPRs from mammals enabled us to identify critical residues of functional importance and suggested that the plant flavin mononucleotide-binding domain might be more flexible than mammalian homologs. In addition, the elucidated structure of SbCPR2b included the first observation of NADP + in a native CPR. Overall, we conclude that the connecting domain of SbCPR2, especially its hinge region, could serve as a target to alter biomass composition in bioenergy and forage sorghums through protein engineering.
- Department of Chemistry, Washington State University, Pullman, Washington, USA.
Organizational Affiliation: