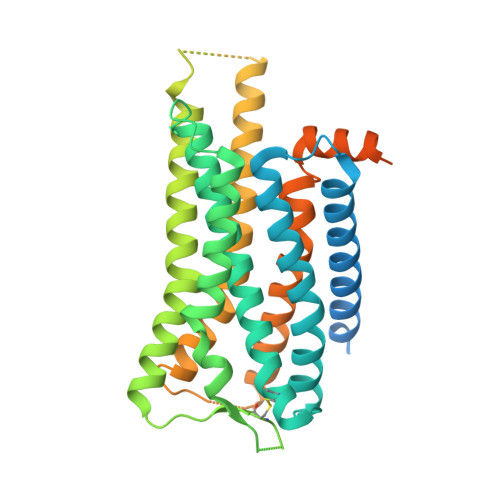
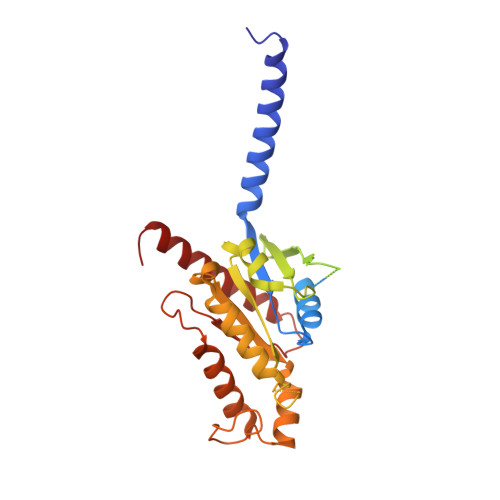
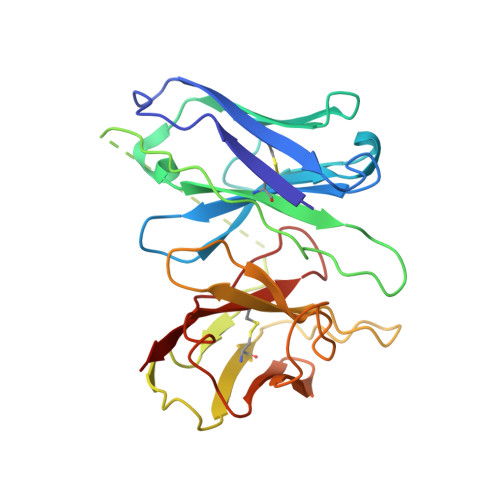
Molecular mechanism of the wake-promoting agent TAK-925.
Yin, J., Kang, Y., McGrath, A.P., Chapman, K., Sjodt, M., Kimura, E., Okabe, A., Koike, T., Miyanohana, Y., Shimizu, Y., Rallabandi, R., Lian, P., Bai, X., Flinspach, M., De Brabander, J.K., Rosenbaum, D.M.(2022) Nat Commun 13: 2902-2902
- PubMed: 35614071
- DOI: https://doi.org/10.1038/s41467-022-30601-3
- Primary Citation of Related Structures:
7SQO, 7SR8 - PubMed Abstract:
The OX 2 orexin receptor (OX 2 R) is a highly expressed G protein-coupled receptor (GPCR) in the brain that regulates wakefulness and circadian rhythms in humans. Antagonism of OX 2 R is a proven therapeutic strategy for insomnia drugs, and agonism of OX 2 R is a potentially powerful approach for narcolepsy type 1, which is characterized by the death of orexinergic neurons. Until recently, agonism of OX 2 R had been considered 'undruggable.' We harness cryo-electron microscopy of OX 2 R-G protein complexes to determine how the first clinically tested OX 2 R agonist TAK-925 can activate OX 2 R in a highly selective manner. Two structures of TAK-925-bound OX 2 R with either a G q mimetic or G i reveal that TAK-925 binds at the same site occupied by antagonists, yet interacts with the transmembrane helices to trigger activating microswitches. Our structural and mutagenesis data show that TAK-925's selectivity is mediated by subtle differences between OX 1 and OX 2 receptor subtypes at the orthosteric pocket. Finally, differences in the polarity of interactions at the G protein binding interfaces help to rationalize OX 2 R's coupling selectivity for G q signaling. The mechanisms of TAK-925's binding, activation, and selectivity presented herein will aid in understanding the efficacy of small molecule OX 2 R agonists for narcolepsy and other circadian disorders.
- Department of Biophysics, The University of Texas Southwestern Medical Center, 6001 Forest Park Road, Dallas, TX, 75390, USA.
Organizational Affiliation: