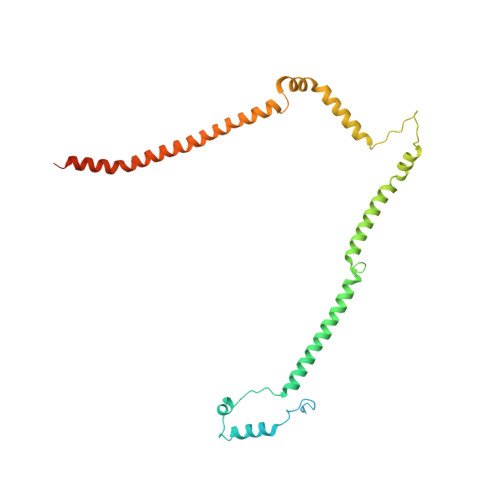
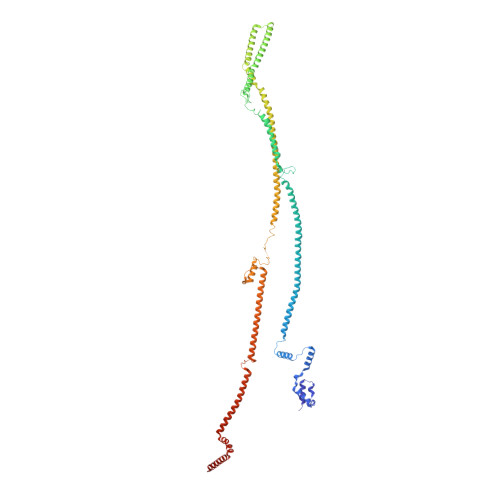
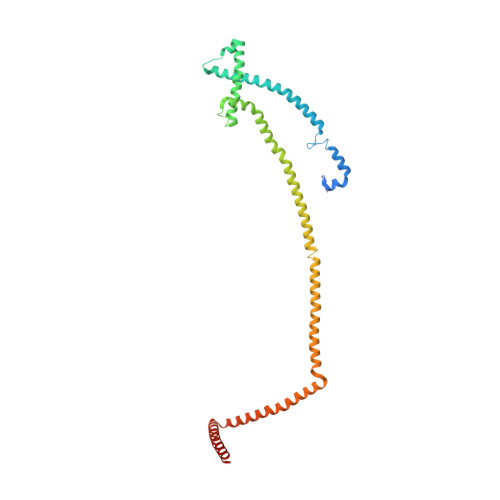
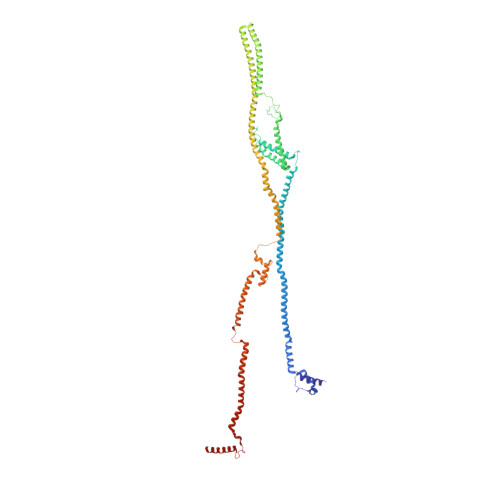
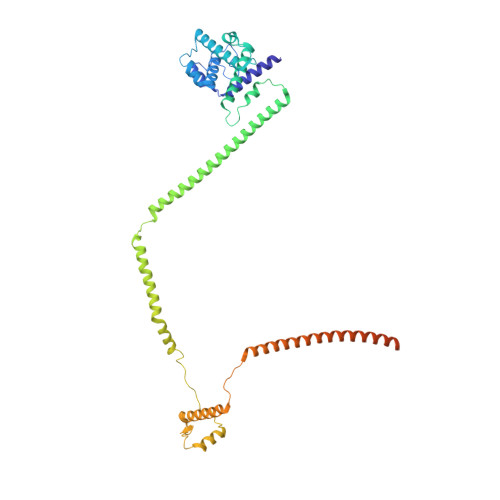
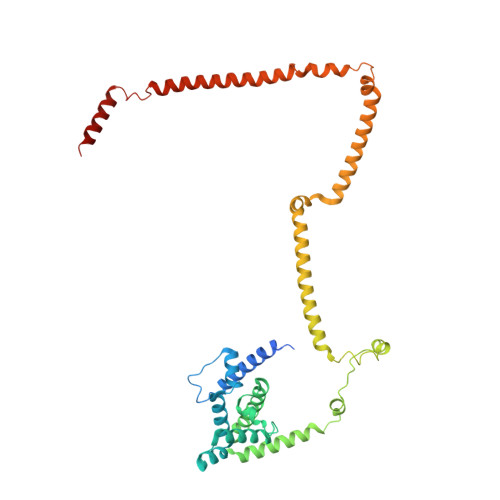
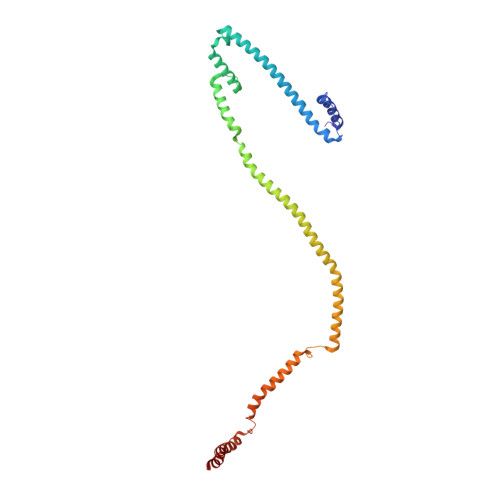Molecular architecture of the augmin complex.
Gabel, C.A., Li, Z., DeMarco, A.G., Zhang, Z., Yang, J., Hall, M.C., Barford, D., Chang, L.(2022) Nat Commun 13: 5449-5449
- PubMed: 36114186
- DOI: https://doi.org/10.1038/s41467-022-33227-7
- Primary Citation of Related Structures:
7SQK - PubMed Abstract:
Accurate segregation of chromosomes during mitosis depends on the correct assembly of the mitotic spindle, a bipolar structure composed mainly of microtubules. The augmin complex, or homologous to augmin subunits (HAUS) complex, is an eight-subunit protein complex required for building robust mitotic spindles in metazoa. Augmin increases microtubule density within the spindle by recruiting the γ-tubulin ring complex (γ-TuRC) to pre-existing microtubules and nucleating branching microtubules. Here, we elucidate the molecular architecture of augmin by single particle cryo-electron microscopy (cryo-EM), computational methods, and crosslinking mass spectrometry (CLMS). Augmin's highly flexible structure contains a V-shaped head and a filamentous tail, with the head existing in either extended or contracted conformational states. Our work highlights how cryo-EM, complemented by computational advances and CLMS, can elucidate the structure of a challenging protein complex and provides insights into the function of augmin in mediating microtubule branching nucleation.
- Department of Biological Sciences, Purdue University, West Lafayette, IN, 47907, USA.
Organizational Affiliation: