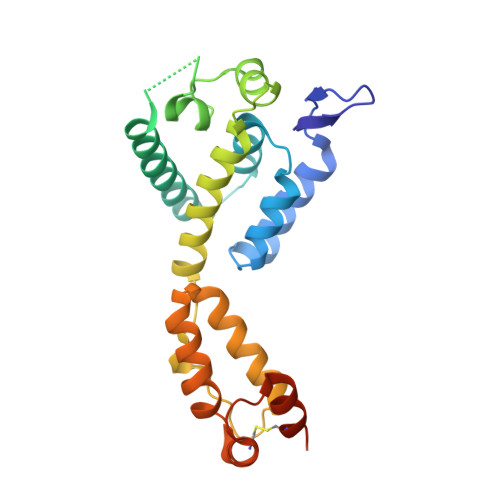
Prion-like low complexity regions enable avid virus-host interactions during HIV-1 infection.
Wei, G., Iqbal, N., Courouble, V.V., Francis, A.C., Singh, P.K., Hudait, A., Annamalai, A.S., Bester, S., Huang, S.W., Shkriabai, N., Briganti, L., Haney, R., KewalRamani, V.N., Voth, G.A., Engelman, A.N., Melikyan, G.B., Griffin, P.R., Asturias, F., Kvaratskhelia, M.(2022) Nat Commun 13: 5879-5879
- PubMed: 36202818
- DOI: https://doi.org/10.1038/s41467-022-33662-6
- Primary Citation of Related Structures:
7SNQ - PubMed Abstract:
Cellular proteins CPSF6, NUP153 and SEC24C play crucial roles in HIV-1 infection. While weak interactions of short phenylalanine-glycine (FG) containing peptides with isolated capsid hexamers have been characterized, how these cellular factors functionally engage with biologically relevant mature HIV-1 capsid lattices is unknown. Here we show that prion-like low complexity regions (LCRs) enable avid CPSF6, NUP153 and SEC24C binding to capsid lattices. Structural studies revealed that multivalent CPSF6 assembly is mediated by LCR-LCR interactions, which are templated by binding of CPSF6 FG peptides to a subset of hydrophobic capsid pockets positioned along adjoining hexamers. In infected cells, avid CPSF6 LCR-mediated binding to HIV-1 cores is essential for functional virus-host interactions. The investigational drug lenacapavir accesses unoccupied hydrophobic pockets in the complex to potently impair HIV-1 inside the nucleus without displacing the tightly bound cellular cofactor from virus cores. These results establish previously undescribed mechanisms of virus-host interactions and antiviral action.
- Division of Infectious Diseases, Anschutz Medical Campus, University of Colorado School of Medicine, Aurora, CO, 80045, USA.
Organizational Affiliation: