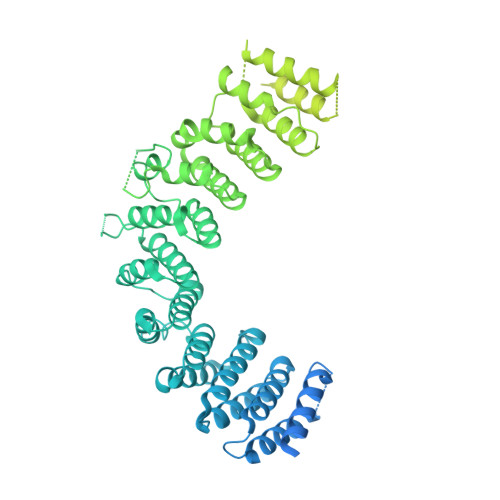
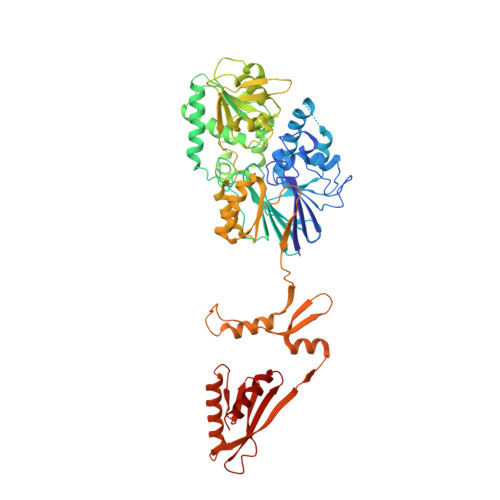
Inositol hexakisphosphate is required for Integrator function.
Lin, M.H., Jensen, M.K., Elrod, N.D., Huang, K.L., Welle, K.A., Wagner, E.J., Tong, L.(2022) Nat Commun 13: 5742-5742
- PubMed: 36180473
- DOI: https://doi.org/10.1038/s41467-022-33506-3
- Primary Citation of Related Structures:
7SN8 - PubMed Abstract:
Integrator is a multi-subunit protein complex associated with RNA polymerase II (Pol II), with critical roles in noncoding RNA 3'-end processing and transcription attenuation of a broad collection of mRNAs. IntS11 is the endonuclease for RNA cleavage, as a part of the IntS4-IntS9-IntS11 Integrator cleavage module (ICM). Here we report a cryo-EM structure of the Drosophila ICM, at 2.74 Å resolution, revealing stable association of an inositol hexakisphosphate (IP 6 ) molecule. The IP 6 binding site is located in a highly electropositive pocket at an interface among all three subunits of ICM, 55 Å away from the IntS11 active site and generally conserved in other ICMs. We also confirmed IP 6 association with the same site in human ICM. IP 6 binding is not detected in ICM samples harboring mutations in this binding site. Such mutations or disruption of IP 6 biosynthesis significantly reduced Integrator function in snRNA 3'-end processing and mRNA transcription attenuation. Our structural and functional studies reveal that IP 6 is required for Integrator function in Drosophila, humans, and likely other organisms.
- Department of Biological Sciences, Columbia University, New York, NY, 10027, USA.
Organizational Affiliation: