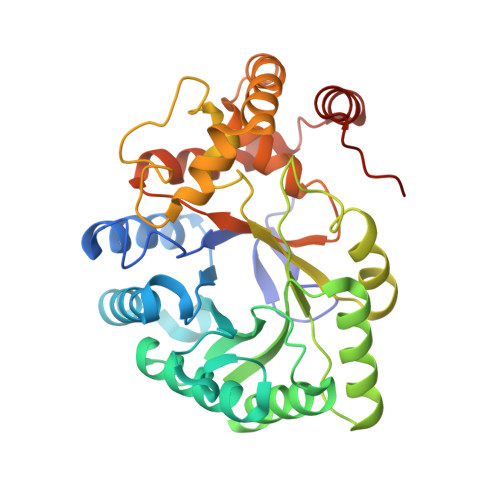
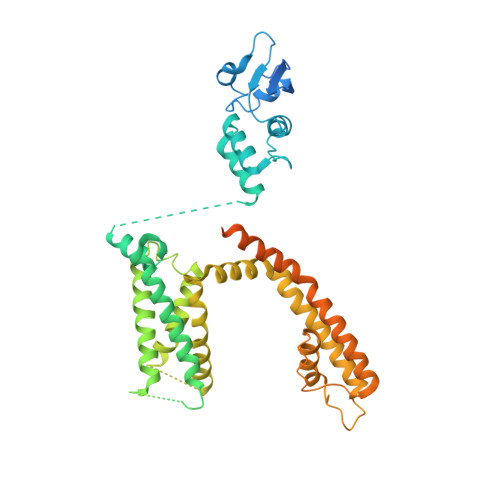
Structural basis for C-type inactivation in a Shaker family voltage-gated K + channel.
Reddi, R., Matulef, K., Riederer, E.A., Whorton, M.R., Valiyaveetil, F.I.(2022) Sci Adv 8: eabm8804-eabm8804
- PubMed: 35452285
- DOI: https://doi.org/10.1126/sciadv.abm8804
- Primary Citation of Related Structures:
7SIT, 7SIZ - PubMed Abstract:
C-type inactivation is a process by which ion flux through a voltage-gated K + (K v ) channel is regulated at the selectivity filter. While prior studies have indicated that C-type inactivation involves structural changes at the selectivity filter, the nature of the changes has not been resolved. Here, we report the crystal structure of the K v 1.2 channel in a C-type inactivated state. The structure shows that C-type inactivation involves changes in the selectivity filter that disrupt the outer two ion binding sites in the filter. The changes at the selectivity filter propagate to the extracellular mouth and the turret regions of the channel pore. The structural changes observed are consistent with the functional hallmarks of C-type inactivation. This study highlights the intricate interplay between K + occupancy at the ion binding sites and the interactions of the selectivity filter in determining the balance between the conductive and the inactivated conformations of the filter.
- Program in Chemical Biology, Department of Chemical Physiology and Biochemistry, Oregon Health & Science University, 3181 SW Sam Jackson Park Rd, Portland, OR 97239, USA.
Organizational Affiliation: