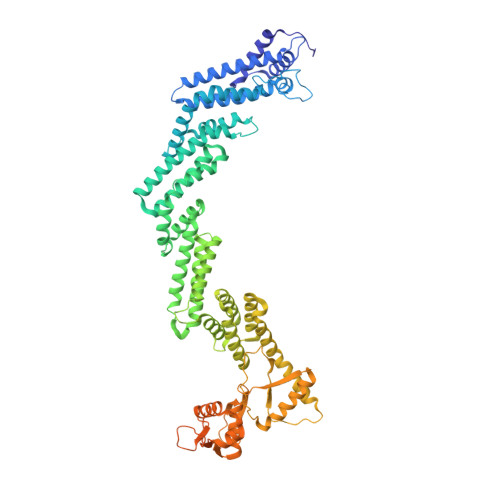
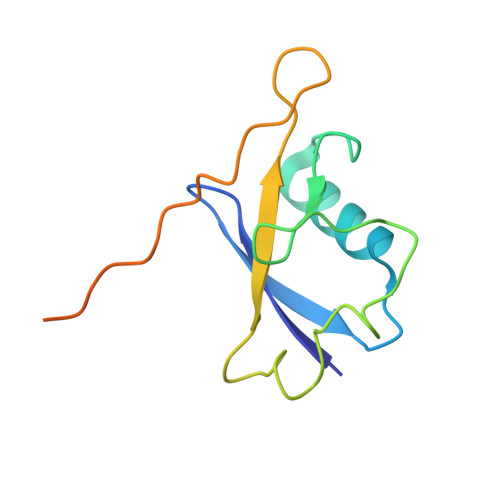
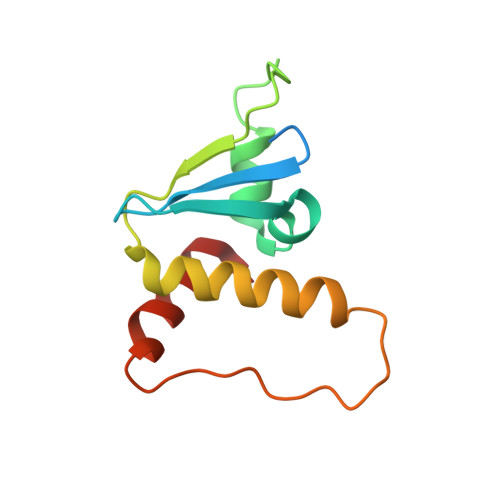
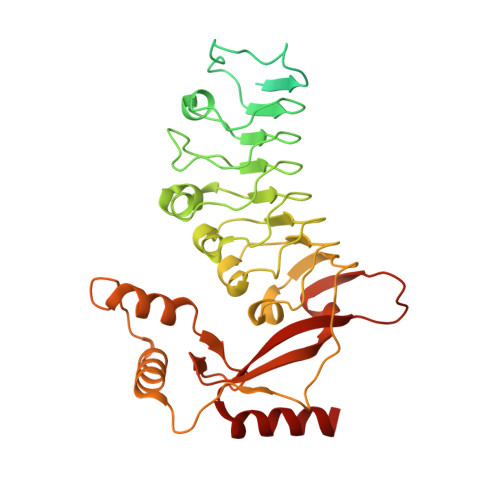
Structure of CRL2Lrr1, the E3 ubiquitin ligase that promotes DNA replication termination in vertebrates.
Zhou, H., Zaher, M.S., Walter, J.C., Brown, A.(2021) Nucleic Acids Res 49: 13194-13206
- PubMed: 34850944
- DOI: https://doi.org/10.1093/nar/gkab1174
- Primary Citation of Related Structures:
7SHK, 7SHL - PubMed Abstract:
When vertebrate replisomes from neighboring origins converge, the Mcm7 subunit of the replicative helicase, CMG, is ubiquitylated by the E3 ubiquitin ligase, CRL2Lrr1. Polyubiquitylated CMG is then disassembled by the p97 ATPase, leading to replication termination. To avoid premature replisome disassembly, CRL2Lrr1 is only recruited to CMGs after they converge, but the underlying mechanism is unclear. Here, we use cryogenic electron microscopy to determine structures of recombinant Xenopus laevis CRL2Lrr1 with and without neddylation. The structures reveal that CRL2Lrr1 adopts an unusually open architecture, in which the putative substrate-recognition subunit, Lrr1, is located far from the catalytic module that catalyzes ubiquitin transfer. We further demonstrate that a predicted, flexible pleckstrin homology domain at the N-terminus of Lrr1 is essential to target CRL2Lrr1 to terminated CMGs. We propose a hypothetical model that explains how CRL2Lrr1's catalytic module is positioned next to the ubiquitylation site on Mcm7, and why CRL2Lrr1 binds CMG only after replisomes converge.
- Department of Biological Chemistry and Molecular Pharmacology, Blavatnik Institute, Harvard Medical School, 240 Longwood Avenue, Boston, MA 02115, USA.
Organizational Affiliation: