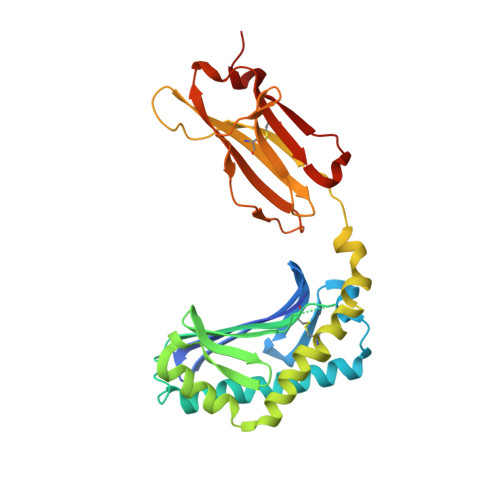
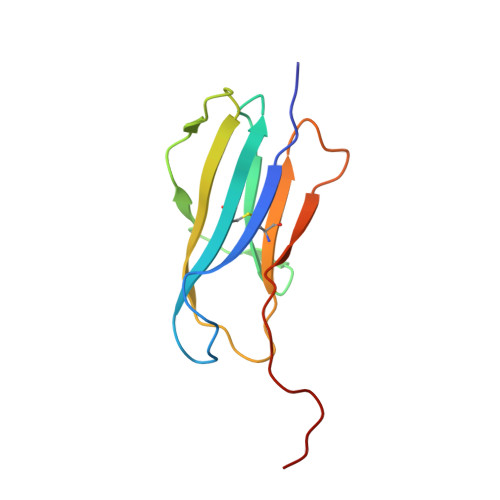
Staphylococcal phosphatidylglycerol antigens activate human T cells via CD1a.
Monnot, G.C., Wegrecki, M., Cheng, T.Y., Chen, Y.L., Sallee, B.N., Chakravarthy, R., Karantza, I.M., Tin, S.Y., Khaleel, A.E., Monga, I., Uwakwe, L.N., Tillman, A., Cheng, B., Youssef, S., Ng, S.W., Shahine, A., Garcia-Vilas, J.A., Uhlemann, A.C., Bordone, L.A., Han, A., Rohde, C.H., Ogg, G., Moody, D.B., Rossjohn, J., de Jong, A.(2023) Nat Immunol 24: 110-122
- PubMed: 36550321
- DOI: https://doi.org/10.1038/s41590-022-01375-z
- Primary Citation of Related Structures:
7SH4 - PubMed Abstract:
Expressed on epidermal Langerhans cells, CD1a presents a range of self-lipid antigens found within the skin; however, the extent to which CD1a presents microbial ligands from bacteria colonizing the skin is unclear. Here we identified CD1a-dependent T cell responses to phosphatidylglycerol (PG), a ubiquitous bacterial membrane phospholipid, as well as to lysylPG, a modified PG, present in several Gram-positive bacteria and highly abundant in Staphylococcus aureus. The crystal structure of the CD1a-PG complex showed that the acyl chains were buried within the A'- and F'-pockets of CD1a, while the phosphoglycerol headgroup remained solvent exposed in the F'-portal and was available for T cell receptor contact. Using lysylPG and PG-loaded CD1a tetramers, we identified T cells in peripheral blood and in skin that respond to these lipids in a dose-dependent manner. Tetramer + CD4 + T cell lines secreted type 2 helper T cell cytokines in response to phosphatidylglycerols as well as to co-cultures of CD1a + dendritic cells and Staphylococcus bacteria. The expansion in patients with atopic dermatitis of CD4 + CD1a-(lysyl)PG tetramer + T cells suggests a response to lipids made by bacteria associated with atopic dermatitis and provides a link supporting involvement of PG-based lipid-activated T cells in atopic dermatitis pathogenesis.
- Department of Dermatology, Columbia University Irving Medical Center, New York, NY, USA.
Organizational Affiliation: