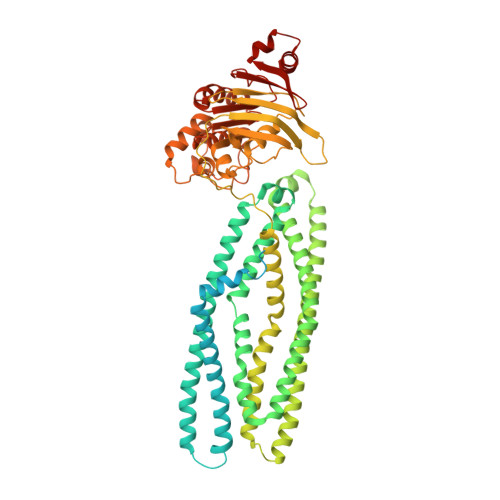The hemolysin A secretion system is a multi-engine pump containing three ABC transporters.
Zhao, H., Lee, J., Chen, J.(2022) Cell 185: 3329-3340.e13
- PubMed: 36055198
- DOI: https://doi.org/10.1016/j.cell.2022.07.017
- Primary Citation of Related Structures:
7SGR, 8DCK - PubMed Abstract:
Type 1 secretion systems (T1SSs) are widespread in pathogenic Gram-negative bacteria, extruding protein substrates following synthesis of the entire polypeptide. The Escherichia coli hemolysin A secretion system has long been considered a prototype in structural and mechanistic studies of T1SSs. Three membrane proteins-an inner membrane ABC transporter HlyB, an adaptor protein HlyD, and an outer membrane porin TolC-are required for secretion. However, the stoichiometry and structure of the complex are unknown. Here, cryo-electron microscopy (cryo-EM) structures determined in two conformations reveal that the inner membrane complex is a hetero-dodecameric assembly comprising three HlyB homodimers and six HlyD subunits. Functional studies indicate that oligomerization of HlyB and HlyD is essential for protein secretion and that polypeptides translocate through a canonical ABC transporter pathway in HlyB. Our data suggest that T1SSs entail three ABC transporters, one that functions as a protein channel and two that allosterically power the translocation process.
- Laboratory of Membrane Biology and Biophysics, The Rockefeller University, New York, NY 10065, USA.
Organizational Affiliation: