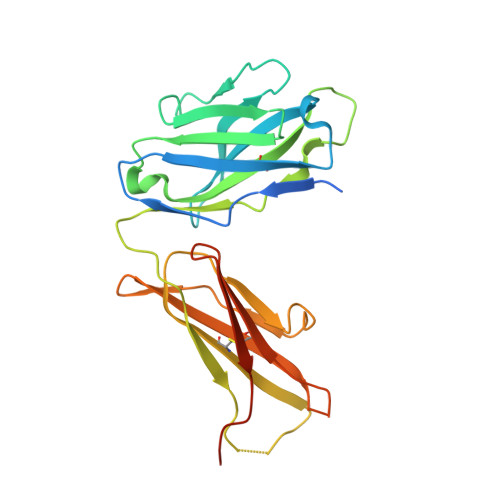
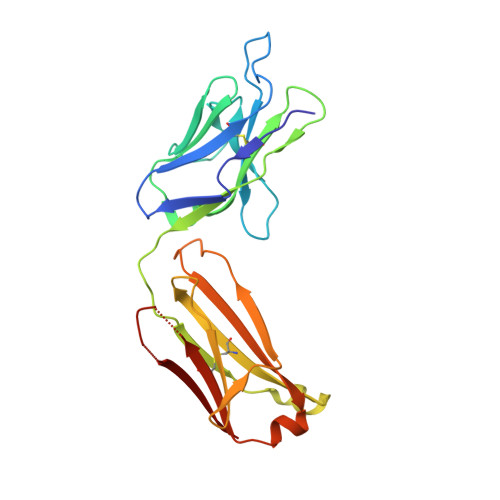
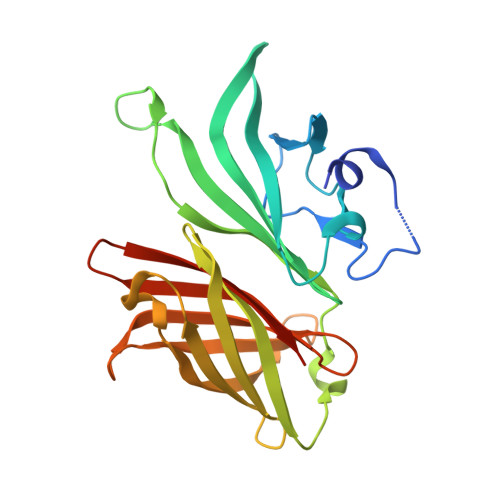
Active Learning for Rapid Design: An iterative AI approach for accelerated vaccine design that combines active machine learning and high-throughput experimental evaluation
Chesterman, C., Desautels, T., Sierra, L.J., Arrildt, K., Zemla, A., Lau, E.Y., Sundaram, S., Laliberte, J., Chen, L., Ruby, A., Mednikov, M., Bertholet, S., Yu, D., Luisi, K., Malito, E., Mallet, C., Bottomley, M.J., van den Berg, R.A., Faissol, D.To be published.