Structure of the K. lactis telomerase RNA binding domain
Skordalakes, E., Tzfati, Y.(2022) Int J Mol Sci
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2022) Int J Mol Sci
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Telomerase reverse transcriptase | 237 | Kluyveromyces lactis | Mutation(s): 0 Gene Names: KLLA0_C18381g EC: 2.7.7.49 | 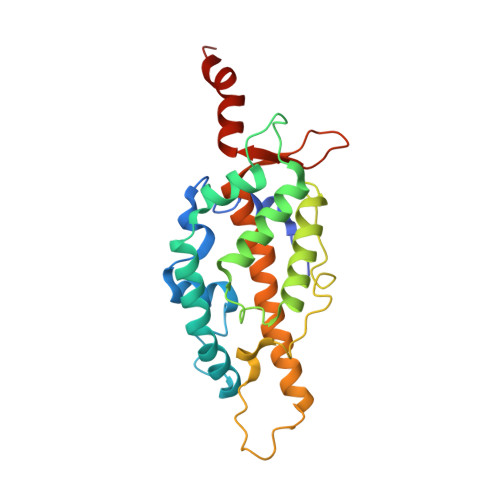 | |
UniProt | |||||
Find proteins for Q6CSS0 (Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37)) Explore Q6CSS0 Go to UniProtKB: Q6CSS0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6CSS0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 140.37 | α = 90 |
| b = 85 | β = 106.11 |
| c = 48.42 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| XSCALE | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Cancer Institute (NIH/NCI) | United States | -- |