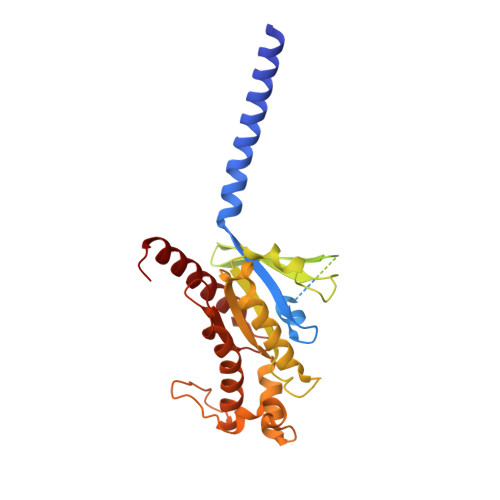
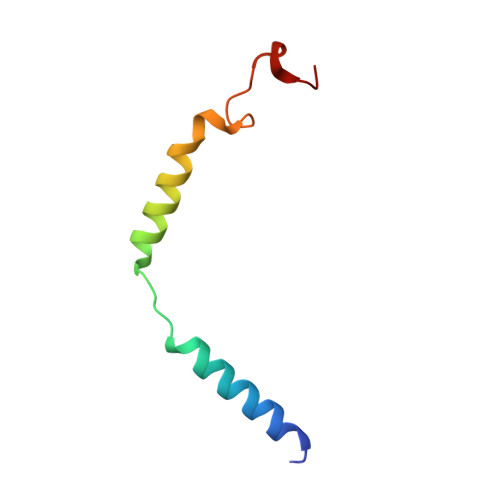
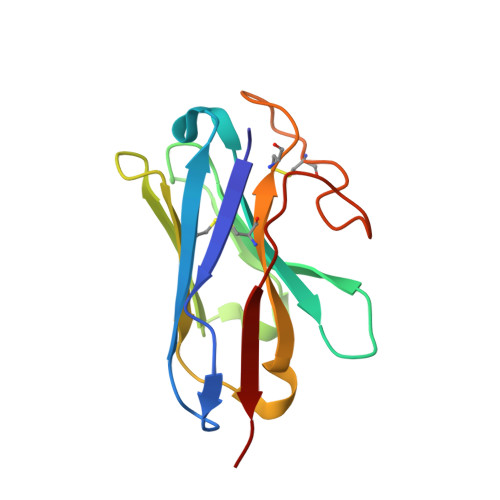
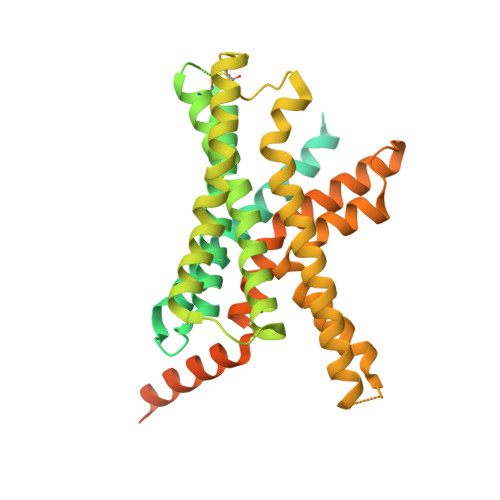
Structural and functional diversity among agonist-bound states of the GLP-1 receptor.
Cary, B.P., Deganutti, G., Zhao, P., Truong, T.T., Piper, S.J., Liu, X., Belousoff, M.J., Danev, R., Sexton, P.M., Wootten, D., Gellman, S.H.(2022) Nat Chem Biol 18: 256-263
- PubMed: 34937906
- DOI: https://doi.org/10.1038/s41589-021-00945-w
- Primary Citation of Related Structures:
7S1M, 7S3I - PubMed Abstract:
Recent advances in G-protein-coupled receptor (GPCR) structural elucidation have strengthened previous hypotheses that multidimensional signal propagation mediated by these receptors depends, in part, on their conformational mobility; however, the relationship between receptor function and static structures is inherently uncertain. Here, we examine the contribution of peptide agonist conformational plasticity to activation of the glucagon-like peptide 1 receptor (GLP-1R), an important clinical target. We use variants of the peptides GLP-1 and exendin-4 (Ex4) to explore the interplay between helical propensity near the agonist N terminus and the ability to bind to and activate the receptor. Cryo-EM analysis of a complex involving an Ex4 analog, the GLP-1R and G s heterotrimer revealed two receptor conformers with distinct modes of peptide-receptor engagement. Our functional and structural data, along with molecular dynamics (MD) simulations, suggest that receptor conformational dynamics associated with flexibility of the peptide N-terminal activation domain may be a key determinant of agonist efficacy.
- Department of Chemistry, University of Wisconsin-Madison, Madison, WI, USA.
Organizational Affiliation: