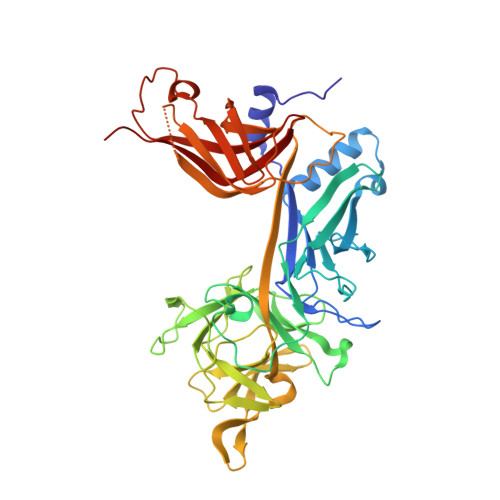
The structure, binding and function of a Notch transcription complex involving RBPJ and the epigenetic reader protein L3MBTL3.
Hall, D., Giaimo, B.D., Park, S.S., Hemmer, W., Friedrich, T., Ferrante, F., Bartkuhn, M., Yuan, Z., Oswald, F., Borggrefe, T., Rual, J.F., Kovall, R.A.(2022) Nucleic Acids Res 50: 13083-13099
- PubMed: 36477367
- DOI: https://doi.org/10.1093/nar/gkac1137
- Primary Citation of Related Structures:
7RTE, 7RTI - PubMed Abstract:
The Notch pathway transmits signals between neighboring cells to elicit downstream transcriptional programs. Notch is a major regulator of cell fate specification, proliferation, and apoptosis, such that aberrant signaling leads to a pleiotropy of human diseases, including developmental disorders and cancers. The pathway signals through the transcription factor CSL (RBPJ in mammals), which forms an activation complex with the intracellular domain of the Notch receptor and the coactivator Mastermind. CSL can also function as a transcriptional repressor by forming complexes with one of several different corepressor proteins, such as FHL1 or SHARP in mammals and Hairless in Drosophila. Recently, we identified L3MBTL3 as a bona fide RBPJ-binding corepressor that recruits the repressive lysine demethylase LSD1/KDM1A to Notch target genes. Here, we define the RBPJ-interacting domain of L3MBTL3 and report the 2.06 Å crystal structure of the RBPJ-L3MBTL3-DNA complex. The structure reveals that L3MBTL3 interacts with RBPJ via an unusual binding motif compared to other RBPJ binding partners, which we comprehensively analyze with a series of structure-based mutants. We also show that these disruptive mutations affect RBPJ and L3MBTL3 function in cells, providing further insights into Notch mediated transcriptional regulation.
- University of Cincinnati College of Medicine, Department of Molecular Genetics, Biochemistry and Microbiology, Cincinnati, OH, USA.
Organizational Affiliation: