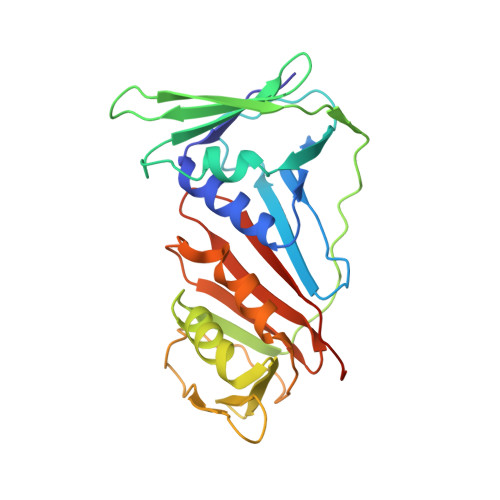
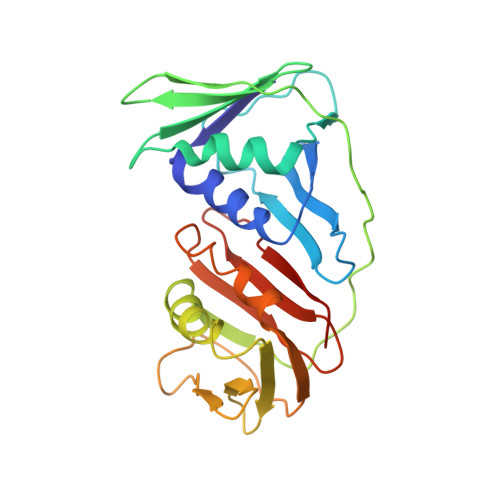
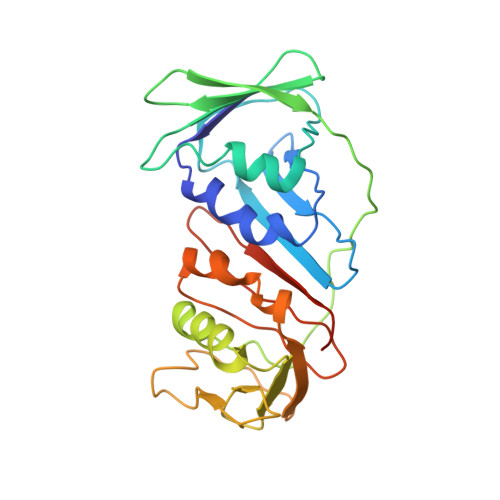
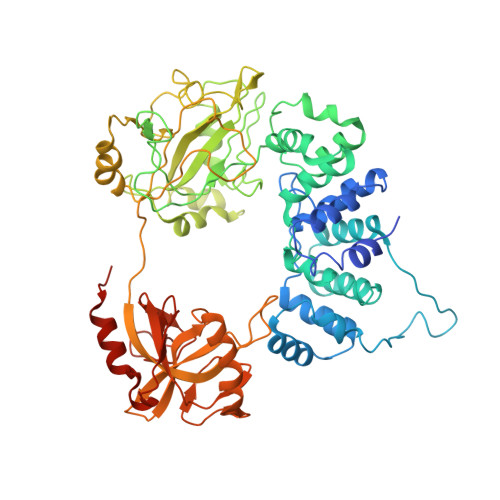
Cryo-EM structures and biochemical insights into heterotrimeric PCNA regulation of DNA ligase.
Sverzhinsky, A., Tomkinson, A.E., Pascal, J.M.(2022) Structure 30: 371
- PubMed: 34838188
- DOI: https://doi.org/10.1016/j.str.2021.11.002
- Primary Citation of Related Structures:
7RPO, 7RPW, 7RPX - PubMed Abstract:
DNA ligases act in the final step of many DNA repair pathways and are commonly regulated by the DNA sliding clamp proliferating cell nuclear antigen (PCNA), but there are limited insights into the physical basis for this regulation. Here, we use single-particle cryoelectron microscopy (cryo-EM) to analyze an archaeal DNA ligase and heterotrimeric PCNA in complex with a single-strand DNA break. The cryo-EM structures highlight a continuous DNA-binding surface formed between DNA ligase and PCNA that supports the distorted conformation of the DNA break undergoing repair and contributes to PCNA stimulation of DNA ligation. DNA ligase is conformationally flexible within the complex, with its domains fully ordered only when encircling the repaired DNA to form a stacked ring structure with PCNA. The structures highlight DNA ligase structural transitions while docked on PCNA, changes in DNA conformation during ligation, and the potential for DNA ligase domains to regulate PCNA accessibility to other repair factors.
- Department of Biochemistry and Molecular Medicine, Faculty of Medicine, Université de Montréal, Québec H3T 1J4, Canada.
Organizational Affiliation: