A periplasmic cinched protein is required for siderophore secretion and virulence of Mycobacterium tuberculosis.
Zhang, L., Kent, J.E., Whitaker, M., Young, D.C., Herrmann, D., Aleshin, A.E., Ko, Y.H., Cingolani, G., Saad, J.S., Moody, D.B., Marassi, F.M., Ehrt, S., Niederweis, M.(2022) Nat Commun 13: 2255-2255
- PubMed: 35474308
- DOI: https://doi.org/10.1038/s41467-022-29873-6
- Primary Citation of Related Structures:
7REF - PubMed Abstract:
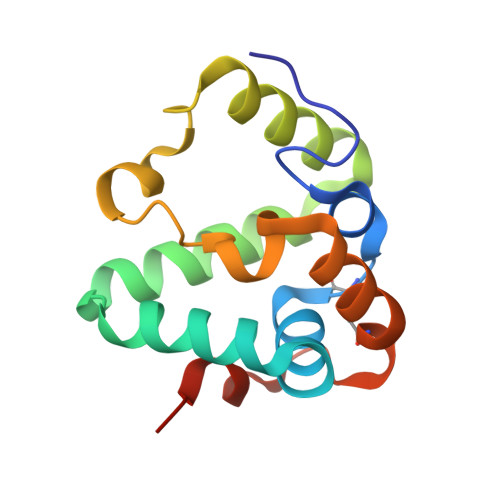
Iron is essential for growth of Mycobacterium tuberculosis, the causative agent of tuberculosis. To acquire iron from the host, M. tuberculosis uses the siderophores called mycobactins and carboxymycobactins. Here, we show that the rv0455c gene is essential for M. tuberculosis to grow in low-iron medium and that secretion of both mycobactins and carboxymycobactins is drastically reduced in the rv0455c deletion mutant. Both water-soluble and membrane-anchored Rv0455c are functional in siderophore secretion, supporting an intracellular role. Lack of Rv0455c results in siderophore toxicity, a phenotype observed for other siderophore secretion mutants, and severely impairs replication of M. tuberculosis in mice, demonstrating the importance of Rv0455c and siderophore secretion during disease. The crystal structure of a Rv0455c homolog reveals a novel protein fold consisting of a helical bundle with a 'cinch' formed by an essential intramolecular disulfide bond. These findings advance our understanding of the distinct M. tuberculosis siderophore secretion system.
- Department of Microbiology, University of Alabama at Birmingham, Birmingham, AL, 35294, USA.
Organizational Affiliation: