Validation and promise of a TCR mimic antibody for cancer immunotherapy of hepatocellular carcinoma.
Liu, C., Liu, H., Dasgupta, M., Hellman, L.M., Zhang, X., Qu, K., Xue, H., Wang, Y., Fan, F., Chang, Q., Yu, D., Ge, L., Zhang, Y., Cui, Z., Zhang, P., Heller, B., Zhang, H., Shi, B., Baker, B.M., Liu, C.(2022) Sci Rep 12: 12068-12068
- PubMed: 35840635
- DOI: https://doi.org/10.1038/s41598-022-15946-5
- Primary Citation of Related Structures:
7RE7, 7RE8, 7RE9 - PubMed Abstract:
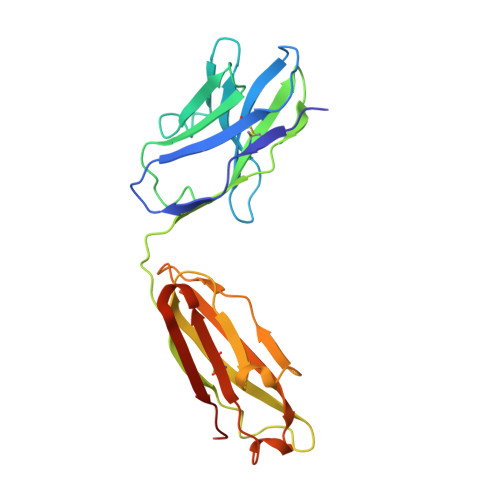
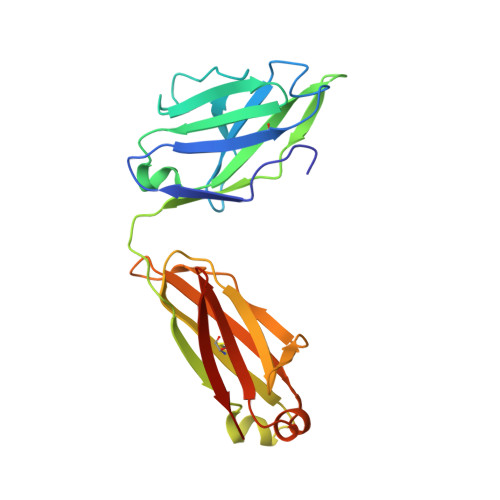
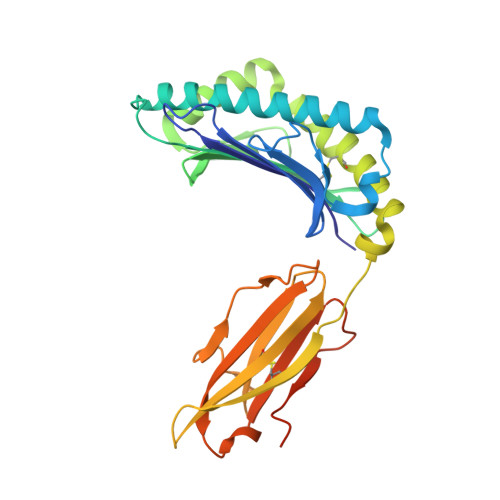
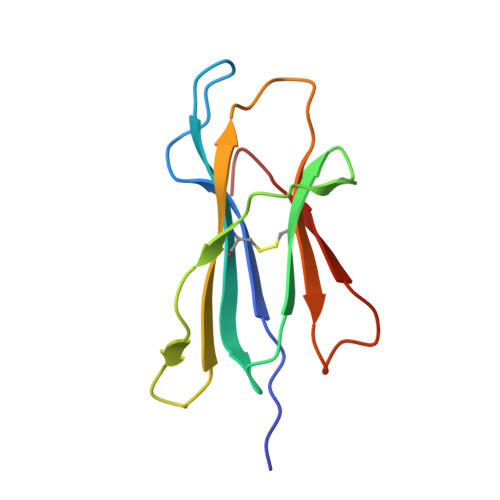
Monoclonal antibodies are at the vanguard of the most promising cancer treatments. Whereas traditional therapeutic antibodies have been limited to extracellular antigens, T cell receptor mimic (TCRm) antibodies can target intracellular antigens presented by cell surface major histocompatibility complex (MHC) proteins. TCRm antibodies can therefore target a repertoire of otherwise undruggable cancer antigens. However, the consequences of off-target peptide/MHC recognition with engineered T cell therapies are severe, and thus there are significant safety concerns with TCRm antibodies. Here we explored the specificity and safety profile of a new TCRm-based T cell therapy for hepatocellular carcinoma (HCC), a solid tumor for which no effective treatment exists. We targeted an alpha-fetoprotein peptide presented by HLA-A*02 with a highly specific TCRm, which crystallographic structural analysis showed binds directly over the HLA protein and interfaces with the full length of the peptide. We fused the TCRm to the γ and δ subunits of a TCR, producing a signaling AbTCR construct. This was combined with an scFv/CD28 co-stimulatory molecule targeting glypican-3 for increased efficacy towards tumor cells. This AbTCR + co-stimulatory T cell therapy showed potent activity against AFP-positive cancer cell lines in vitro and an in an in vivo model and undetectable activity against AFP-negative cells. In an in-human safety assessment, no significant adverse events or cytokine release syndrome were observed and evidence of efficacy was seen. Remarkably, one patient with metastatic HCC achieved a complete remission after nine months and ultimately qualified for a liver transplant.
- The First Affiliated Hospital of Xi'an Jiaotong University, Xi'an, Shaanxi Province, China.
Organizational Affiliation: