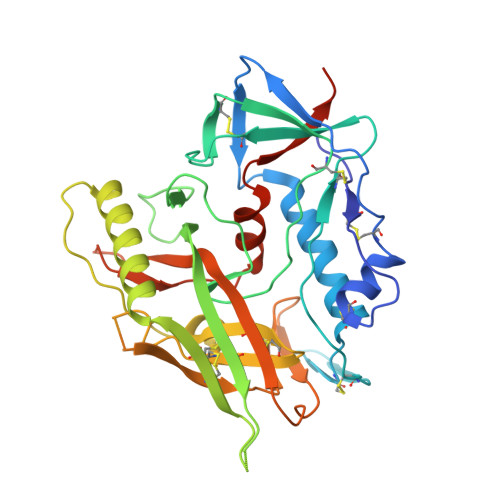
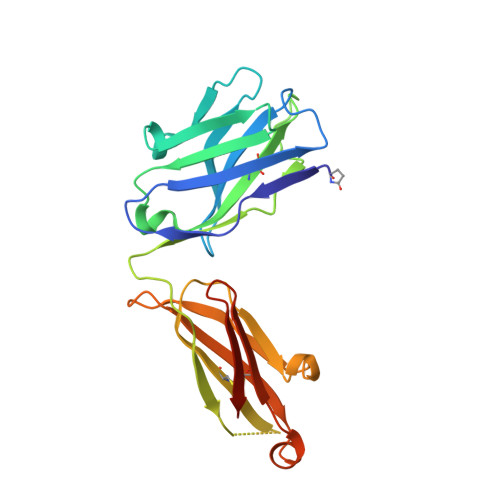
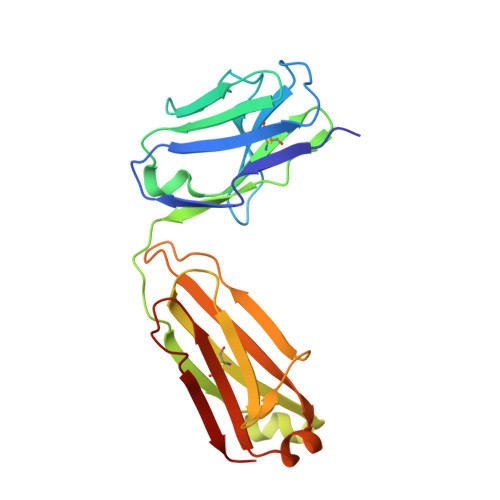
Characterization of a vaccine-elicited human antibody with sequence homology to VRC01-class antibodies that binds the C1C2 gp120 domain.
Gray, M.D., Feng, J., Weidle, C.E., Cohen, K.W., Ballweber-Fleming, L., MacCamy, A.J., Huynh, C.N., Trichka, J.J., Montefiori, D., Ferrari, G., Pancera, M., McElrath, M.J., Stamatatos, L.(2022) Sci Adv 8: eabm3948-eabm3948
- PubMed: 35507661
- DOI: https://doi.org/10.1126/sciadv.abm3948
- Primary Citation of Related Structures:
7RDW - PubMed Abstract:
Broadly HIV-1-neutralizing VRC01-class antibodies bind the CD4-binding site of Env and contain V H 1-2*02-derived heavy chains paired with light chains expressing five-amino acid-long CDRL3s. Their unmutated germline forms do not recognize HIV-1 Env, and their lack of elicitation in human clinical trials could be due to the absence of activation of the corresponding naïve B cells by the vaccine immunogens. To address this point, we examined Env-specific B cell receptor sequences from participants in the HVTN 100 clinical trial. Of all the sequences analyzed, only one displayed homology to VRC01-class antibodies, but the corresponding antibody (FH1) recognized the C1C2 gp120 domain. For FH1 to switch epitope recognition to the CD4-binding site, alterations in the CDRH3 and CDRL3 were necessary. Only germ line-targeting Env immunogens efficiently activated VRC01 B cells, even in the presence of FH1 B cells. Our findings support the use of these immunogens to activate VRC01 B cells in humans.
- Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA.
Organizational Affiliation: