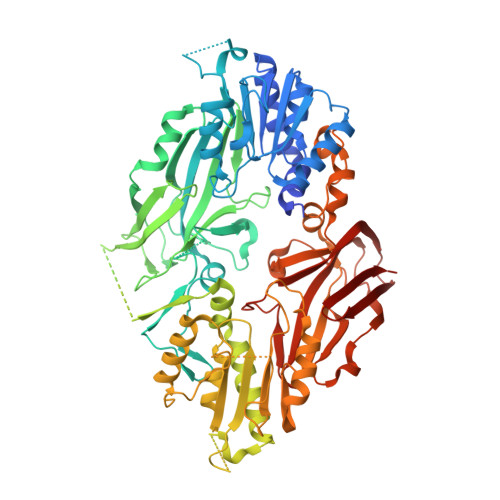
Co-crystal structure of human PRMT9 in complex with MT556 inhibitor
Zeng, H., Dong, A., Hutchinson, A., Seitova, A., Li, Y., Gao, Y.D., Schneider, S., Siliphaivanh, P., Sloman, D., Nicholson, B., Fischer, C., Hicks, J., Brown, P.J., Arrowsmith, C.H., Edwards, A.M., Halabelian, L., Structural Genomics Consortium (SGC)To be published.