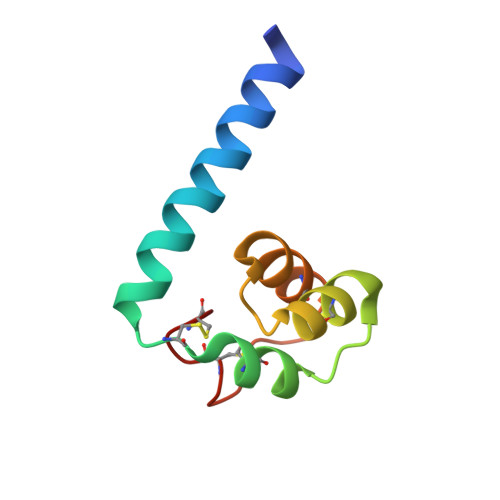
Cbp1, a fungal virulence factor under positive selection, forms an effector complex that drives macrophage lysis.
Azimova, D., Herrera, N., Duvenage, L., Voorhies, M., Rodriguez, R.A., English, B.C., Hoving, J.C., Rosenberg, O., Sil, A.(2022) PLoS Pathog 18: e1010417-e1010417
- PubMed: 35731824
- DOI: https://doi.org/10.1371/journal.ppat.1010417
- Primary Citation of Related Structures:
7R6U, 7R79 - PubMed Abstract:
Intracellular pathogens secrete effectors to manipulate their host cells. Histoplasma capsulatum (Hc) is a fungal intracellular pathogen of humans that grows in a yeast form in the host. Hc yeasts are phagocytosed by macrophages, where fungal intracellular replication precedes macrophage lysis. The most abundant virulence factor secreted by Hc yeast cells is Calcium Binding Protein 1 (Cbp1), which is absolutely required for macrophage lysis. Here we take an evolutionary, structural, and cell biological approach to understand Cbp1 function. We find that Cbp1 is present only in the genomes of closely related dimorphic fungal species of the Ajellomycetaceae family that lead primarily intracellular lifestyles in their mammalian hosts (Histoplasma, Paracoccidioides, and Emergomyces), but not conserved in the extracellular fungal pathogen Blastomyces dermatitidis. We observe a high rate of fixation of non-synonymous substitutions in the Cbp1 coding sequences, indicating that Cbp1 is under positive selection. We determine the de novo structures of Hc H88 Cbp1 and the Paracoccidioides americana (Pb03) Cbp1, revealing a novel "binocular" fold consisting of a helical dimer arrangement wherein two helices from each monomer contribute to a four-helix bundle. In contrast to Pb03 Cbp1, we show that Emergomyces Cbp1 orthologs are unable to stimulate macrophage lysis when expressed in the Hc cbp1 mutant. Consistent with this result, we find that wild-type Emergomyces africanus yeast are able to grow within primary macrophages but are incapable of lysing them. Finally, we use subcellular fractionation of infected macrophages and indirect immunofluorescence to show that Cbp1 localizes to the macrophage cytosol during Hc infection, making this the first instance of a phagosomal human fungal pathogen directing an effector into the cytosol of the host cell. We additionally show that Cbp1 forms a complex with Yps-3, another known Hc virulence factor that accesses the cytosol. Taken together, these data imply that Cbp1 is a fungal virulence factor under positive selection that localizes to the cytosol to trigger host cell lysis.
- University of California San Francisco, San Francisco, California, United States of America.
Organizational Affiliation: