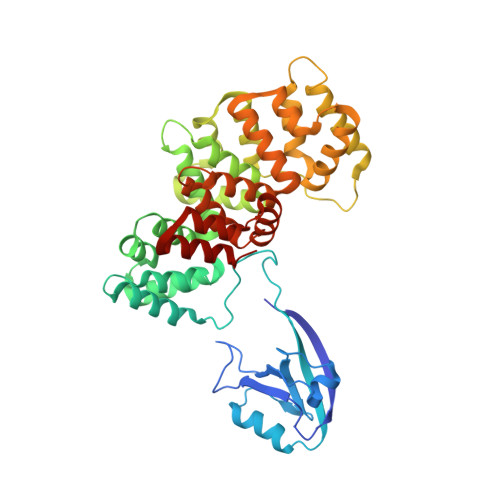
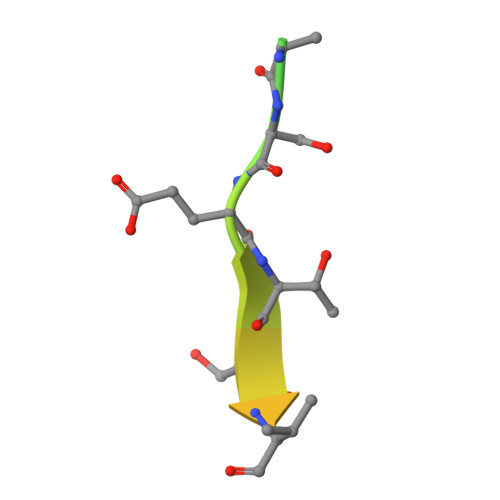
PDZome-wide and structural characterization of the PDZ-binding motif of VANGL2.
Montserrat-Gomez, M., Gogl, G., Carrasco, K., Betzi, S., Durbesson, F., Cousido-Siah, A., Kostmann, C., Essig, D.J., Stromgaard, K., Ostergaard, S., Morelli, X., Trave, G., Vincentelli, R., Bailly, E., Borg, J.P.(2024) Biochim Biophys Acta Proteins Proteom 1872: 140989-140989
- PubMed: 38142947
- DOI: https://doi.org/10.1016/j.bbapap.2023.140989
- Primary Citation of Related Structures:
7R2M, 7R2T - PubMed Abstract:
VANGL2 is a core component of the non-canonical Wnt/Planar Cell Polarity signaling pathway that uses its highly conserved carboxy-terminal type 1 PDZ-binding motif (PBM) to bind a variety of PDZ proteins. In this study, we characterize and quantitatively assess the largest VANGL2 PDZome-binding profile documented so far, using orthogonal methods. The results of our holdup approach support VANGL2 interactions with a large panel of both long-recognized and unprecedented PDZ domains. Truncation and point mutation analyses of the VANGL2 PBM establish that, beyond the strict requirement of the P-0 / V521 and P-2 / T519 amino acids, upstream residues, including E518, Q516 and R514 at, respectively, P-3, P-5 and P-7 further contribute to the robustness of VANGL2 interactions with two distinct PDZ domains, SNX27 and SCRIBBLE-PDZ3. In agreement with these data, incremental amino-terminal deletions of the VANGL2 PBM causes its overall affinity to progressively decline. Moreover, the holdup data establish that the PDZome binding repertoire of VANGL2 starts to diverge significantly with the truncation of E518. A structural analysis of the SYNJ2BP-PDZ/VANGL2 interaction with truncated PBMs identifies a major conformational change in the binding direction of the PBM peptide after the P-2 position. Finally, we report that the PDZome binding profile of VANGL2 is dramatically rearranged upon phosphorylation of S517, T519 and S520. Our crystallographic approach illustrates how SYNJ2BP accommodates a S520-phosphorylated PBM peptide through the ideal positioning of two basic residues, K48 and R86. Altogether our data provides a comprehensive view of the VANGL2 PDZ network and how this network specifically responds to the post-translation modification of distinct PBM residues. These findings should prove useful in guiding future functional and molecular studies of the key PCP component VANGL2.
- Aix Marseille Université, CNRS, INSERM, Institut Paoli-Calmettes, CRCM, Equipe labellisée Ligue 'Cell polarity, cell signaling and cancer', Marseille, France.
Organizational Affiliation: