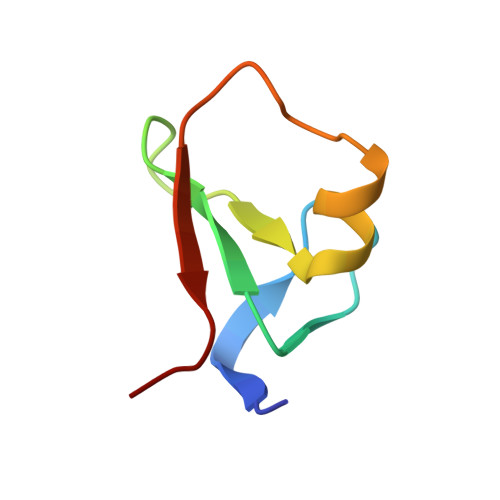
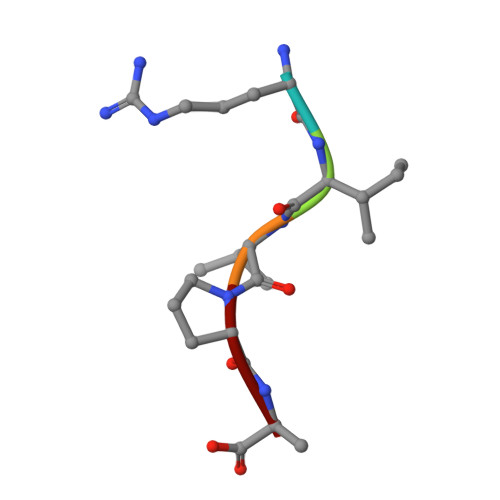
Dusquetide modulates innate immune response through binding to p62.
Zhang, Y., Towers, C.G., Singh, U.K., Liu, J., Hakansson, M., Logan, D.T., Donini, O., Kutateladze, T.G.(2022) Structure 30: 1055
- PubMed: 35640615
- DOI: https://doi.org/10.1016/j.str.2022.05.003
- Primary Citation of Related Structures:
7R1O - PubMed Abstract:
SQSTM1/p62 is an autophagic receptor that plays a major role in mediating stress and innate immune responses. Preclinical studies identified p62 as a target of the prototype innate defense regulator (IDR); however, the molecular mechanism of this process remains unclear. Here, we describe the structural basis and biological consequences of the interaction of p62 with the next generation of IDRs, dusquetide. Both electrostatic and hydrophobic contacts drive the formation of the complex between dusquetide and the ZZ domain of p62. We show that dusquetide penetrates the cell membrane and associates with p62 in vivo. Dusquetide binding modulates the p62-RIP1 complex, increases p38 phosphorylation, and enhances CEBP/B expression without activating autophagy. Our findings provide molecular details underlying the IDR action that may help in the development of new strategies to pharmacologically target p62.
- Department of Pharmacology, University of Colorado School of Medicine, Aurora, CO 80045, USA; Department of Biochemistry, Case Western Reserve University, Cleveland, OH 44106, USA. Electronic address: yi.zhang26@case.edu.
Organizational Affiliation: