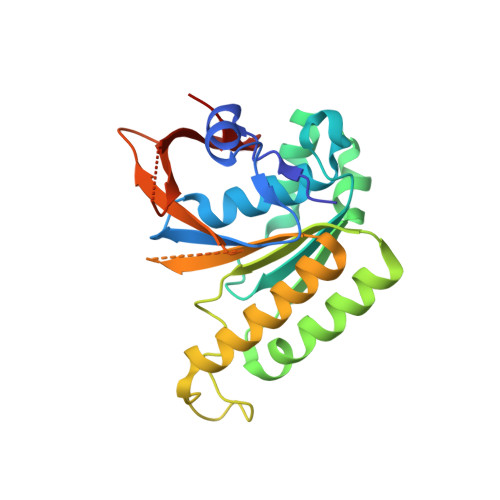
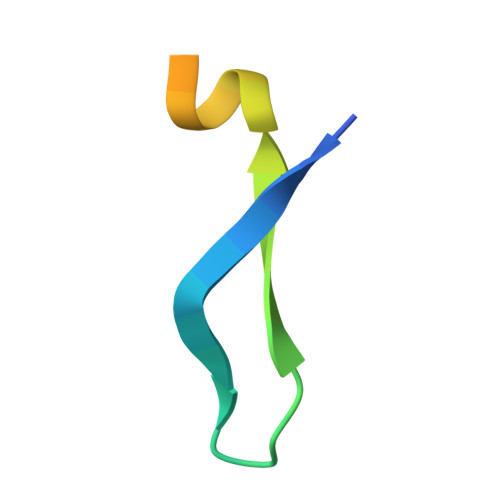
Divergent binding mode for a protozoan BRC repeat to RAD51.
Pantelejevs, T., Hyvonen, M.(2022) Biochem J 479: 1031-1043
- PubMed: 35502837
- DOI: https://doi.org/10.1042/BCJ20220141
- Primary Citation of Related Structures:
7QV8 - PubMed Abstract:
Interaction of BRCA2 through ca. 30 amino acid residue motifs, BRC repeats, with RAD51 is a conserved feature of the double-strand DNA break repair by homologous recombination in eukaryotes. In humans the binding of the eight BRC repeats is defined by two sequence motifs, FxxA and LFDE, interacting with distinct sites on RAD51. Little is known of the interaction of BRC repeats in other species, especially in protozoans, where variable number of BRC repeats are found in BRCA2 proteins. Here, we have studied in detail the interactions of the two BRC repeats in Leishmania infantum BRCA2 with RAD51. We show LiBRC1 is a high-affinity repeat and determine the crystal structure of its complex with LiRAD51. Using truncation mutagenesis of the LiBRC1 repeat, we demonstrate that high affinity binding is maintained in the absence of an LFDE-like motif and suggest compensatory structural features. These observations point towards a divergent evolution of BRC repeats, where a common FxxA-binding ancestor evolved additional contacts for affinity maturation and fine-tuning.
- Department of Biochemistry, University of Cambridge, Cambridge CB2 1GA, U.K.
Organizational Affiliation: