NMR study of the structure and dynamics of the BRCT domain from the kinetochore protein KKT4.
Ludzia, P., Hayashi, H., Robinson, T., Akiyoshi, B., Redfield, C.(2024) Biomol NMR Assign 18: 15-25
- PubMed: 38453826
- DOI: https://doi.org/10.1007/s12104-024-10163-9
- Primary Citation of Related Structures:
7QRO - PubMed Abstract:
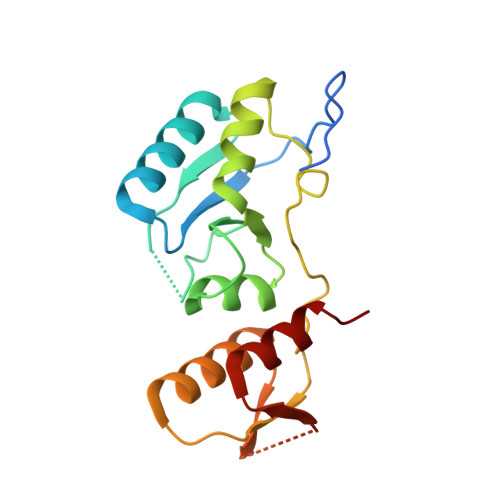
KKT4 is a multi-domain kinetochore protein specific to kinetoplastids, such as Trypanosoma brucei. It lacks significant sequence similarity to known kinetochore proteins in other eukaryotes. Our recent X-ray structure of the C-terminal region of KKT4 shows that it has a tandem BRCT (BRCA1 C Terminus) domain fold with a sulfate ion bound in a typical binding site for a phosphorylated serine or threonine. Here we present the 1 H, 13 C and 15 N resonance assignments for the BRCT domain of KKT4 (KKT4 463-645 ) from T. brucei. We show that the BRCT domain can bind phosphate ions in solution using residues involved in sulfate ion binding in the X-ray structure. We have used these assignments to characterise the secondary structure and backbone dynamics of the BRCT domain in solution. Mutating the residues involved in phosphate ion binding in T. brucei KKT4 BRCT results in growth defects confirming the importance of the BRCT phosphopeptide-binding activity in vivo. These results may facilitate rational drug design efforts in the future to combat diseases caused by kinetoplastid parasites.
- Department of Biochemistry, University of Oxford, South Parks Road, Oxford, OX1 3QU, UK.
Organizational Affiliation: