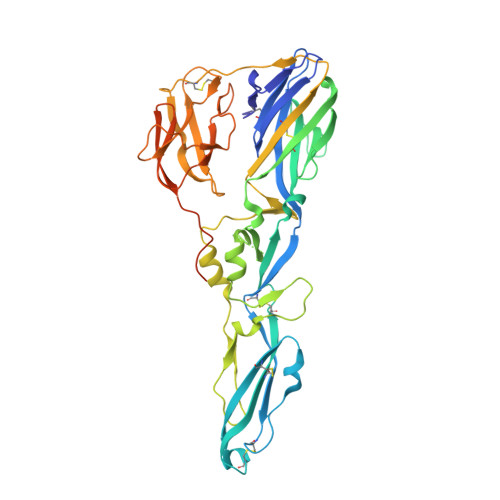
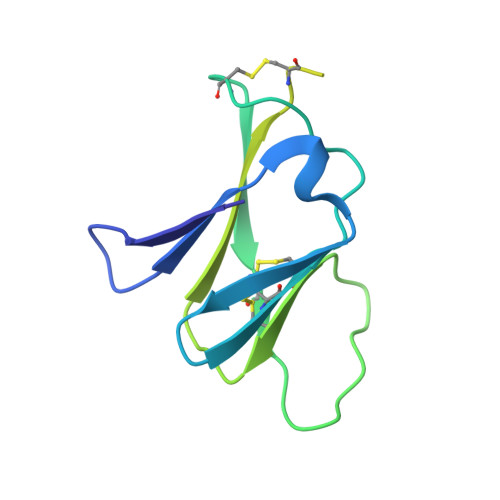
Evolution and activation mechanism of the flavivirus class II membrane-fusion machinery.
Vaney, M.C., Dellarole, M., Duquerroy, S., Medits, I., Tsouchnikas, G., Rouvinski, A., England, P., Stiasny, K., Heinz, F.X., Rey, F.A.(2022) Nat Commun 13: 3718-3718
- PubMed: 35764616
- DOI: https://doi.org/10.1038/s41467-022-31111-y
- Primary Citation of Related Structures:
7QRE, 7QRF, 7QRG - PubMed Abstract:
The flavivirus envelope glycoproteins prM and E drive the assembly of icosahedral, spiky immature particles that bud across the membrane of the endoplasmic reticulum. Maturation into infectious virions in the trans-Golgi network involves an acid-pH-driven rearrangement into smooth particles made of (prM/E) 2 dimers exposing a furin site for prM cleavage into "pr" and "M". Here we show that the prM "pr" moiety derives from an HSP40 cellular chaperonin. Furthermore, the X-ray structure of the tick-borne encephalitis virus (pr/E) 2 dimer at acidic pH reveals the E 150-loop as a hinged-lid that opens at low pH to expose a positively-charged pr-binding pocket at the E dimer interface, inducing (prM/E) 2 dimer formation to generate smooth particles in the Golgi. Furin cleavage is followed by lid-closure upon deprotonation in the neutral-pH extracellular environment, expelling pr while the 150-loop takes the relay in fusion loop protection, thus revealing the elusive flavivirus mechanism of fusion activation.
- Institut Pasteur, Université Paris Cité, CNRS UMR 3569, Unité de Virologie Structurale, Paris, France.
Organizational Affiliation: