A GID E3 ligase assembly ubiquitinates an Rsp5 E3 adaptor and regulates plasma membrane transporters.
Langlois, C.R., Beier, V., Karayel, O., Chrustowicz, J., Sherpa, D., Mann, M., Schulman, B.A.(2022) EMBO Rep 23: e53835-e53835
- PubMed: 35437932
- DOI: https://doi.org/10.15252/embr.202153835
- Primary Citation of Related Structures:
7QQY - PubMed Abstract:
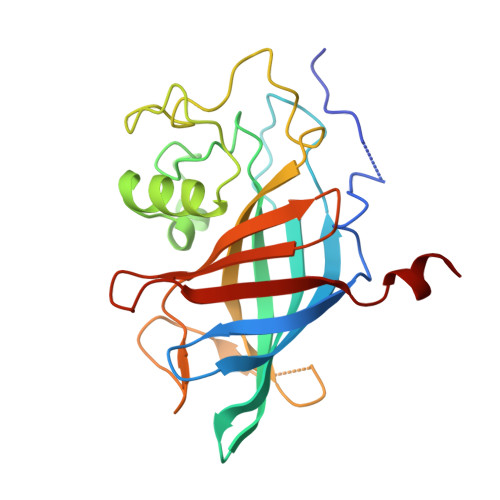
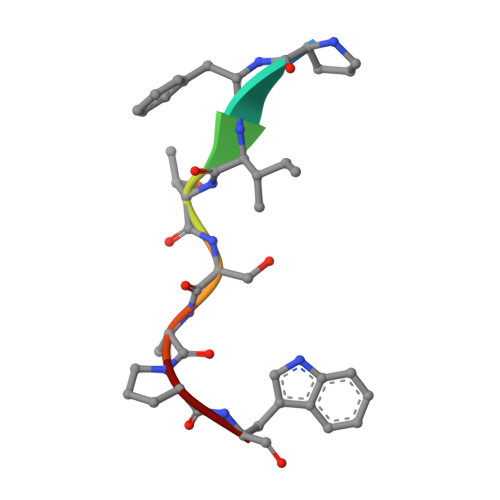
Cells rapidly remodel their proteomes to align their cellular metabolism to environmental conditions. Ubiquitin E3 ligases enable this response, by facilitating rapid and reversible changes to protein stability, localization, or interaction partners. In Saccharomyces cerevisiae, the GID E3 ligase regulates the switch from gluconeogenic to glycolytic conditions through induction and incorporation of the substrate receptor subunit Gid4, which promotes the degradation of gluconeogenic enzymes. Here, we show an alternative substrate receptor, Gid10, which is induced in response to changes in temperature, osmolarity, and nutrient availability, regulates the ART-Rsp5 ubiquitin ligase pathway, a component of plasma membrane quality control. Proteomic studies reveal that the levels of the adaptor protein Art2 are elevated upon GID10 deletion. A crystal structure shows the basis for Gid10-Art2 interactions, and we demonstrate that Gid10 directs a GID E3 ligase complex to ubiquitinate Art2. Our data suggest that the GID E3 ligase affects Art2-dependent amino acid transport. This study reveals GID as a system of E3 ligases with metabolic regulatory functions outside of glycolysis and gluconeogenesis, controlled by distinct stress-specific substrate receptors.
- Department of Molecular Machines and Signaling, Max Planck Institute of Biochemistry, Martinsried, Germany.
Organizational Affiliation: