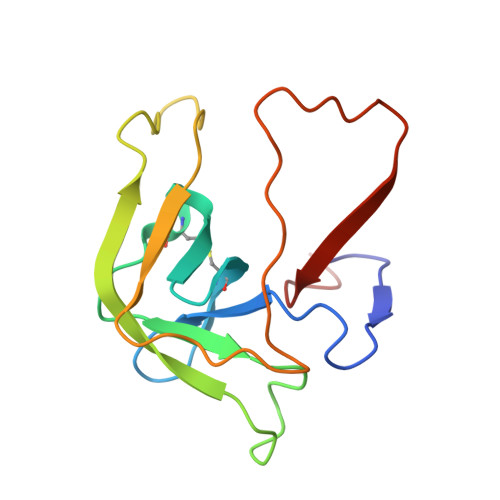
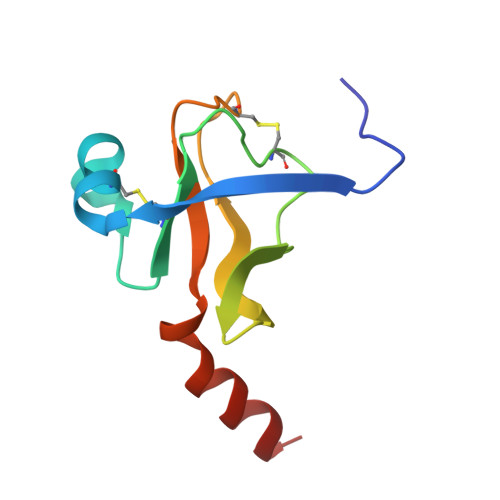
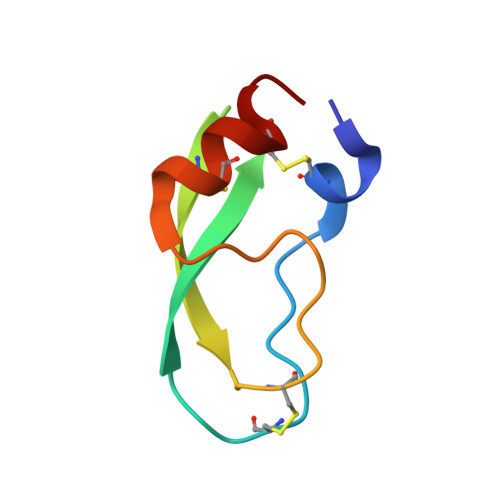
Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Leppkes, J., Dimos, N., Loll, B., Hohmann, T., Dyrks, M., Wieseke, A., Keller, B.G., Koksch, B.(2022) RSC Chem Biol 3: 773-782
- PubMed: 35755190
- DOI: https://doi.org/10.1039/d2cb00018k
- Primary Citation of Related Structures:
7QIQ, 7QIR, 7QIS, 7QIT - PubMed Abstract:
Substituting the P 1 position in bovine pancreatic trypsin inhibitor (BPTI) is known to heavily influence its inhibitory activity towards serine proteases. Side-chain fluorinated aliphatic amino acids have been shown to alter numerous properties of peptides and proteins and thus are of interest in the context of BPTI. In our study, we systematically investigated the site-specific incorporation of non-canonical amino acids into BPTI by microwave-assisted solid-phase peptide synthesis (SPPS). Inhibitor activity of the variants was tested towards the serine protease α-chymotrypsin. We observed enhanced inhibition of two fluorinated BPTIs compared to wild type and hydrocarbon variants. To further investigate the complexes, we performed X-ray structure analysis. Our findings underline the power fluorine offers as a tool in protein engineering to beneficially alter the effects on phenomena as protein-protein interactions.
- Department of Biology, Chemistry and Pharmacy, Institute of Chemistry and Biochemistry, Freie Universität Berlin Arnimallee 20 14195 Berlin Germany beate.koksch@fu-berlin.de.
Organizational Affiliation: