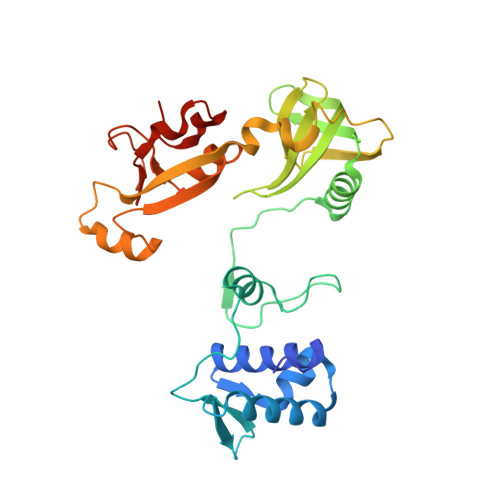
A widespread family of WYL-domain transcriptional regulators co-localizes with diverse phage defence systems and islands.
Picton, D.M., Harling-Lee, J.D., Duffner, S.J., Went, S.C., Morgan, R.D., Hinton, J.C.D., Blower, T.R.(2022) Nucleic Acids Res 50: 5191-5207
- PubMed: 35544231
- DOI: https://doi.org/10.1093/nar/gkac334
- Primary Citation of Related Structures:
7QFZ - PubMed Abstract:
Bacteria are under constant assault by bacteriophages and other mobile genetic elements. As a result, bacteria have evolved a multitude of systems that protect from attack. Genes encoding bacterial defence mechanisms can be clustered into 'defence islands', providing a potentially synergistic level of protection against a wider range of assailants. However, there is a comparative paucity of information on how expression of these defence systems is controlled. Here, we functionally characterize a transcriptional regulator, BrxR, encoded within a recently described phage defence island from a multidrug resistant plasmid of the emerging pathogen Escherichia fergusonii. Using a combination of reporters and electrophoretic mobility shift assays, we discovered that BrxR acts as a repressor. We present the structure of BrxR to 2.15 Å, the first structure of this family of transcription factors, and pinpoint a likely binding site for ligands within the WYL-domain. Bioinformatic analyses demonstrated that BrxR-family homologues are widespread amongst bacteria. About half (48%) of identified BrxR homologues were co-localized with a diverse array of known phage defence systems, either alone or clustered into defence islands. BrxR is a novel regulator that reveals a common mechanism for controlling the expression of the bacterial phage defence arsenal.
- Department of Biosciences, Durham University, Stockton Road, Durham DH1 3LE, UK.
Organizational Affiliation: