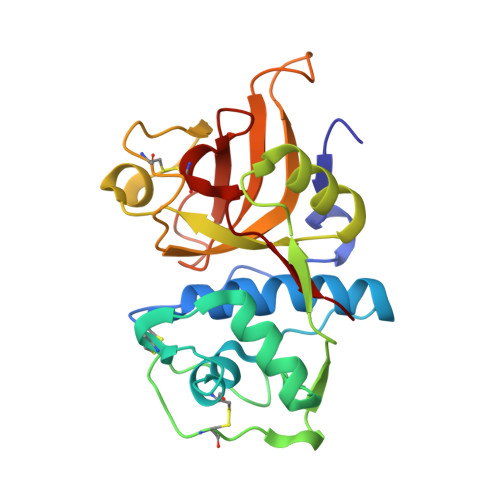
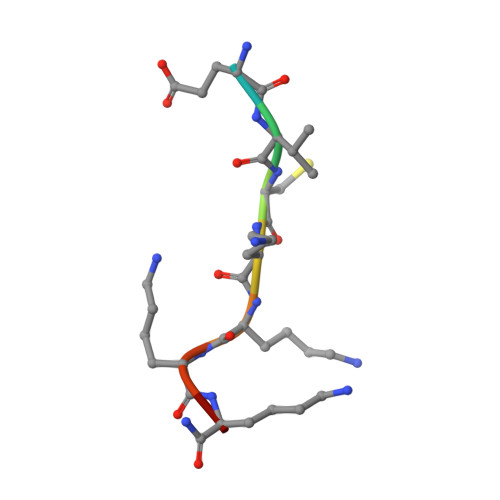
Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Tusar, L., Loboda, J., Impens, F., Sosnowski, P., Van Quickelberghe, E., Vidmar, R., Demol, H., Sedeyn, K., Saelens, X., Vizovisek, M., Mihelic, M., Fonovic, M., Horvat, J., Kosec, G., Turk, B., Gevaert, K., Turk, D.(2023) Commun Biol 6: 450-450
- PubMed: 37095140
- DOI: https://doi.org/10.1038/s42003-023-04772-8
- Primary Citation of Related Structures:
7Q8D, 7Q8F, 7Q8G, 7Q8H, 7Q8I, 7Q8J, 7Q8K, 7Q8L, 7Q8M, 7Q8N, 7Q8O, 7Q8P, 7Q8Q, 7Q9C, 7Q9H, 7QFF, 7QFH, 7QHJ, 7QHK - PubMed Abstract:
Addressing the elusive specificity of cysteine cathepsins, which in contrast to caspases and trypsin-like proteases lack strict specificity determining P1 pocket, calls for innovative approaches. Proteomic analysis of cell lysates with human cathepsins K, V, B, L, S, and F identified 30,000 cleavage sites, which we analyzed by software platform SAPS-ESI (Statistical Approach to Peptidyl Substrate-Enzyme Specific Interactions). SAPS-ESI is used to generate clusters and training sets for support vector machine learning. Cleavage site predictions on the SARS-CoV-2 S protein, confirmed experimentally, expose the most probable first cut under physiological conditions and suggested furin-like behavior of cathepsins. Crystal structure analysis of representative peptides in complex with cathepsin V reveals rigid and flexible sites consistent with analysis of proteomics data by SAPS-ESI that correspond to positions with heterogeneous and homogeneous distribution of residues. Thereby support for design of selective cleavable linkers of drug conjugates and drug discovery studies is provided.
- Jožef Stefan Institute, Department of Biochemistry and Molecular and Structural Biology, Jamova cesta 39, 1000, Ljubljana, Slovenia.
Organizational Affiliation: