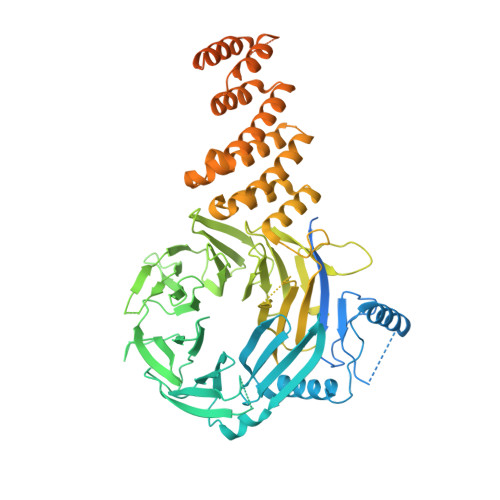
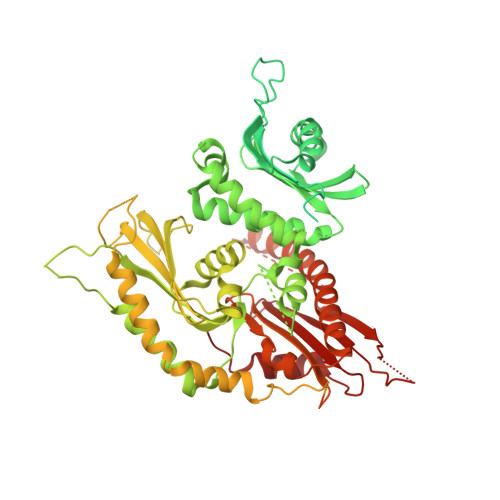
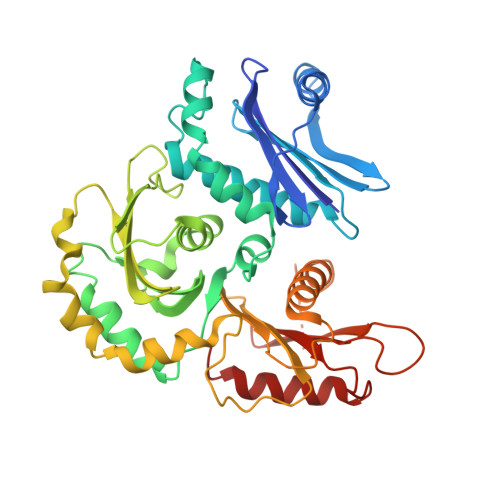
Structure of the ciliogenesis-associated CPLANE complex.
Langousis, G., Cavadini, S., Boegholm, N., Lorentzen, E., Kempf, G., Matthias, P.(2022) Sci Adv 8: eabn0832-eabn0832
- PubMed: 35427153
- DOI: https://doi.org/10.1126/sciadv.abn0832
- Primary Citation of Related Structures:
7Q3D, 7Q3E - PubMed Abstract:
Dysfunctional cilia cause pleiotropic human diseases termed ciliopathies. These hereditary maladies are often caused by defects in cilia assembly, a complex event that is regulated by the ciliogenesis and planar polarity effector (CPLANE) proteins Wdpcp, Inturned, and Fuzzy. CPLANE proteins are essential for building the cilium and are mutated in multiple ciliopathies, yet their structure and molecular functions remain elusive. Here, we show that mammalian CPLANE proteins comprise a bona fide complex and report the near-atomic resolution structures of the human Wdpcp-Inturned-Fuzzy complex and of the mouse Wdpcp-Inturned-Fuzzy complex bound to the small guanosine triphosphatase Rsg1. Notably, the crescent-shaped CPLANE complex binds phospholipids such as phosphatidylinositol 3-phosphate via multiple modules and a CPLANE ciliopathy mutant exhibits aberrant lipid binding. Our study provides critical structural and functional insights into an enigmatic ciliogenesis-associated complex as well as unexpected molecular rationales for ciliopathies.
- Friedrich Miescher Institute for Biomedical Research, 4058 Basel, Switzerland.
Organizational Affiliation: