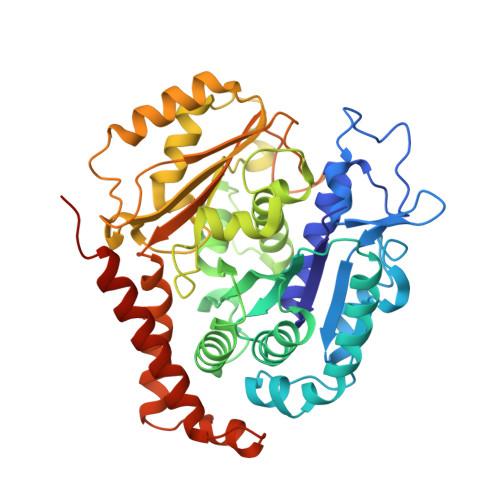
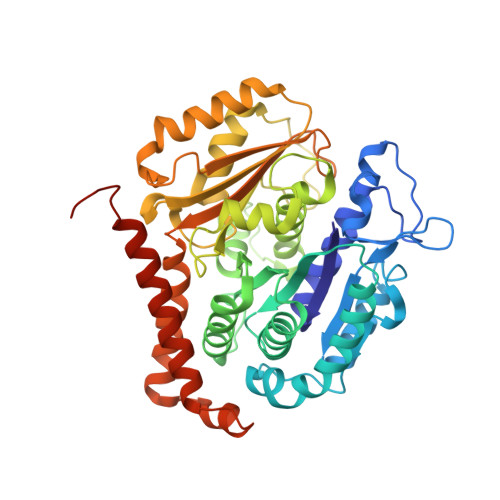
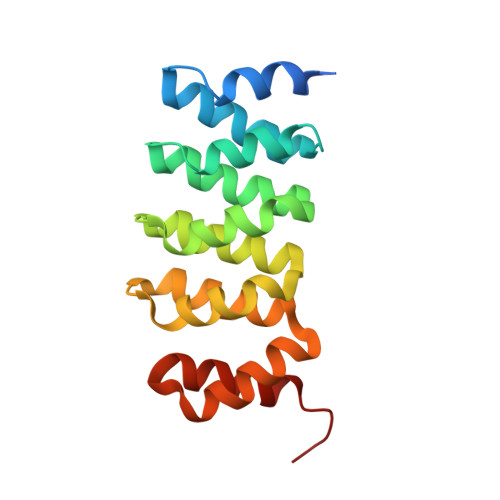
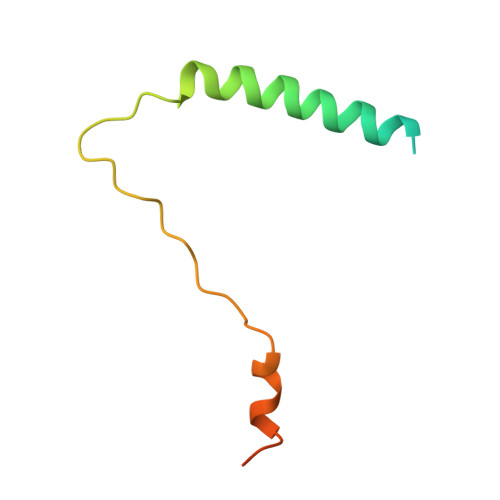
Structural convergence for tubulin binding of CPAP and vinca domain microtubule inhibitors.
Campanacci, V., Urvoas, A., Ammar Khodja, L., Aumont-Nicaise, M., Noiray, M., Lachkar, S., Curmi, P.A., Minard, P., Gigant, B.(2022) Proc Natl Acad Sci U S A 119: e2120098119-e2120098119
- PubMed: 35507869
- DOI: https://doi.org/10.1073/pnas.2120098119
- Primary Citation of Related Structures:
7Q1E, 7Q1F, 7Z0F, 7Z0G - PubMed Abstract:
Microtubule dynamics is regulated by various cellular proteins and perturbed by small-molecule compounds. To what extent the mechanism of the former resembles that of the latter is an open question. We report here structures of tubulin bound to the PN2-3 domain of CPAP, a protein controlling the length of the centrioles. We show that an α-helix of the PN2-3 N-terminal region binds and caps the longitudinal surface of the tubulin β subunit. Moreover, a PN2-3 N-terminal stretch lies in a β-tubulin site also targeted by fungal and bacterial peptide-like inhibitors of the vinca domain, sharing a very similar binding mode with these compounds. Therefore, our results identify several characteristic features of cellular partners that bind to this site and highlight a structural convergence of CPAP with small-molecule inhibitors of microtubule assembly.
- Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198, Gif-sur-Yvette, France.
Organizational Affiliation: