Drosophila melanogaster's Aldehyde Oxidase 1: The First Invertebrate AOX structure
Vilela-Alves, G., Mota, C., Romao, M.J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Aldehyde oxidase 1 | 1,273 | Drosophila melanogaster | Mutation(s): 0 Gene Names: AOX1, CT42272, DmAO1, Dmel\CG18522, LPO, PO, CG18522, Dmel_CG18522 EC: 1.2.3.8 (PDB Primary Data), 1.2.3.1 (UniProt), 1 (UniProt) | 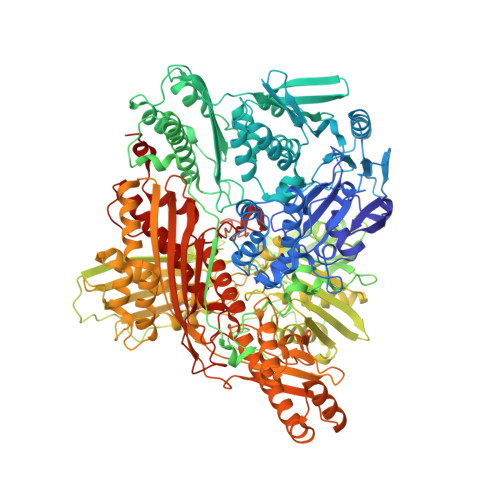 | |
UniProt | |||||
Find proteins for Q9VF53 (Drosophila melanogaster) Explore Q9VF53 Go to UniProtKB: Q9VF53 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9VF53 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FAD (Subject of Investigation/LOI) Query on FAD | DA [auth C], I [auth A], KA [auth D], T [auth B] | FLAVIN-ADENINE DINUCLEOTIDE C27 H33 N9 O15 P2 VWWQXMAJTJZDQX-UYBVJOGSSA-N |  | ||
| MTE (Subject of Investigation/LOI) Query on MTE | BA [auth C], G [auth A], IA [auth D], R [auth B] | PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER C10 H14 N5 O6 P S2 HPEUEJRPDGMIMY-IFQPEPLCSA-N |  | ||
| FES (Subject of Investigation/LOI) Query on FES | AA [auth C] E [auth A] F [auth A] GA [auth D] HA [auth D] | FE2/S2 (INORGANIC) CLUSTER Fe2 S2 NIXDOXVAJZFRNF-UHFFFAOYSA-N |  | ||
| MOS (Subject of Investigation/LOI) Query on MOS | CA [auth C], H [auth A], JA [auth D], S [auth B] | DIOXOTHIOMOLYBDENUM(VI) ION H Mo O2 S BDSRWPHSAKXXRG-UHFFFAOYSA-M |  | ||
| PEG Query on PEG | M [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | EA [auth C] FA [auth C] J [auth A] K [auth A] L [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| NH4 Query on NH4 | NA [auth D] | AMMONIUM ION H4 N QGZKDVFQNNGYKY-UHFFFAOYSA-O |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 144.663 | α = 90 |
| b = 127.914 | β = 110.629 |
| c = 152.547 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Fundacao para a Ciencia e a Tecnologia | Portugal | PTDC/BBB-BEP/1185/2014 |