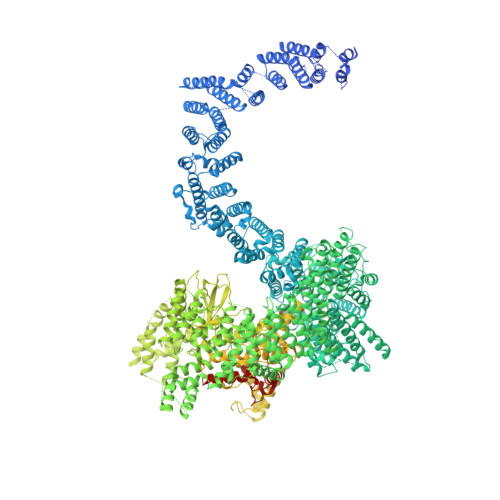
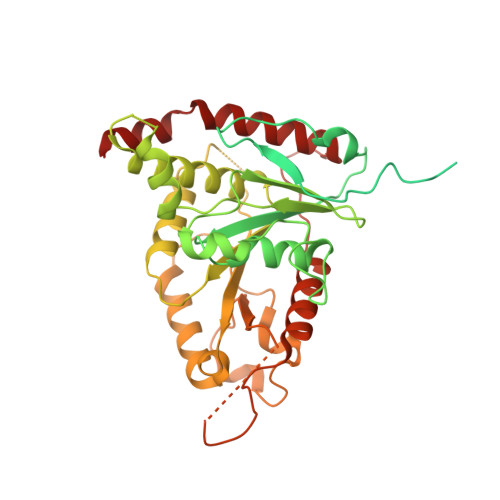
Cryo-EM reconstructions of inhibitor-bound SMG1 kinase reveal an autoinhibitory state dependent on SMG8.
Langer, L.M., Bonneau, F., Gat, Y., Conti, E.(2021) Elife 10
- PubMed: 34698635
- DOI: https://doi.org/10.7554/eLife.72353
- Primary Citation of Related Structures:
7PW4, 7PW5, 7PW6, 7PW7, 7PW8, 7PW9 - PubMed Abstract:
The PI3K-related kinase (PIKK) SMG1 monitors the progression of metazoan nonsense-mediated mRNA decay (NMD) by phosphorylating the RNA helicase UPF1. Previous work has shown that the activity of SMG1 is impaired by small molecule inhibitors, is reduced by the SMG1 interactors SMG8 and SMG9, and is downregulated by the so-called SMG1 insertion domain. However, the molecular basis for this complex regulatory network has remained elusive. Here, we present cryo-electron microscopy reconstructions of human SMG1-9 and SMG1-8-9 complexes bound to either a SMG1 inhibitor or a non-hydrolyzable ATP analog at overall resolutions ranging from 2.8 to 3.6 Å. These structures reveal the basis with which a small molecule inhibitor preferentially targets SMG1 over other PIKKs. By comparison with our previously reported substrate-bound structure (Langer et al.,2020), we show that the SMG1 insertion domain can exert an autoinhibitory function by directly blocking the substrate-binding path as well as overall access to the SMG1 kinase active site. Together with biochemical analysis, our data indicate that SMG1 autoinhibition is stabilized by the presence of SMG8. Our results explain the specific inhibition of SMG1 by an ATP-competitive small molecule, provide insights into regulation of its kinase activity within the NMD pathway, and expand the understanding of PIKK regulatory mechanisms in general.
- Max Planck Institute of Biochemistry, Martinsried, Germany.
Organizational Affiliation: