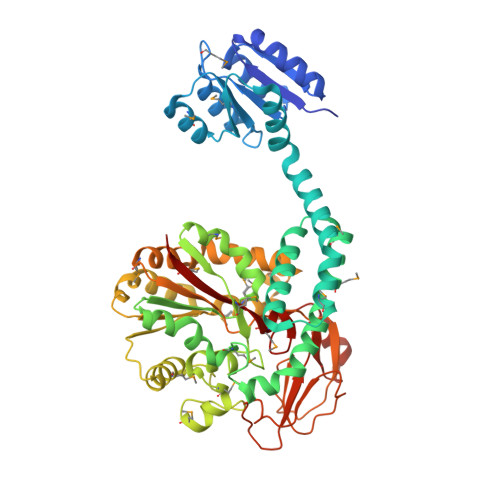
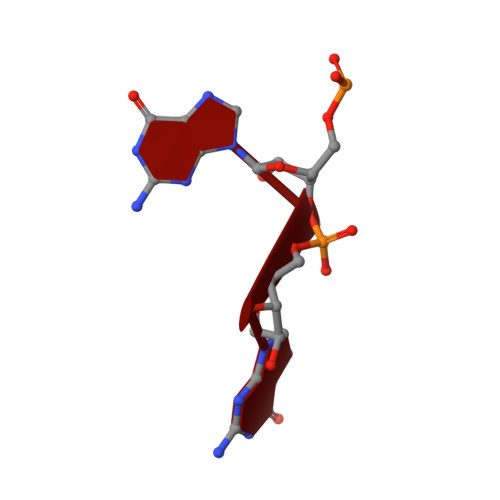
Response regulator PorX coordinates oligonucleotide signalling and gene expression to control the secretion of virulence factors
Schmitz, C., Madej, M., Nowakowska, Z., Cuppari, A., Jacula, A., Ksiazek, M., Mikruta, K., Wisniewski, J., Pudelko-Malik, N., Saran, A., Zeytuni, N., Mlynarz, P., Lamont, R.J., Uson, I., Siksnys, V., Potempa, J., Sola, M.(2022) Nucleic Acids Res 50: 12558-12577